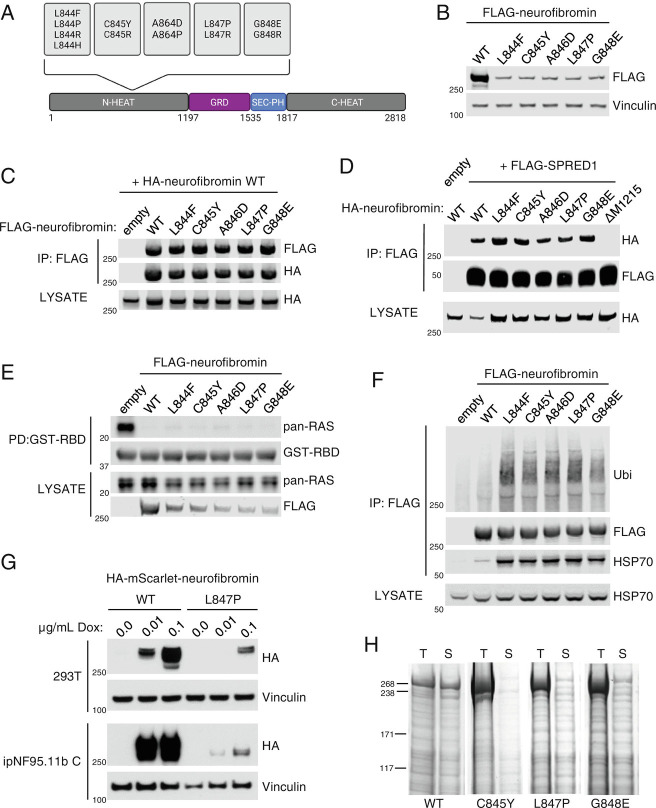
Fig. 1.
844 to 848 neurofibromin mutants are less stable than wild-type protein. (A) Domain structure of neurofibromin showing the location of mutation hotspot. (B) Lysates from 293T NF1−/− cells expressing FLAG-tagged neurofibromin. Expression was analyzed via western blot. (C) FLAG-WT and mutant neurofibromin immunoprecipitates (IPs) from 293T NF1−/− were tested for dimerization with cotransfected HA-WT neurofibromin. The amount of mutant NF1 plasmid was increased to even levels of expression with wild type. (D) HA-tagged WT and mutant neurofibromin coimmunoprecipitated with FLAG-SPRED1. NF1 ΔM1215 is used as a control for the absence of SPRED1 binding. (E) Levels of RAS-GTP by GST-RAF1-RBD (RAS Binding Domain) pulldown from lysates indicate the level of neurofibromin GAP activity in 293T NF1−/−. (F) Ubiquitin (Ubi) and HSP70 were detected on FLAG immunoprecipitants of mutant neurofibromin. (G) Lysates from HA-mScarlet-NF1-transduced 293T and ipNF95.11b C Schwann cells after 48 h Dox treatment at the indicated concentrations. (H) Recombinant mutant neurofibromin proteins are highly insoluble in baculovirus-infected insect cells. Images of Coomassie-stained SDS-PAGE. T = Total, S = Soluble.