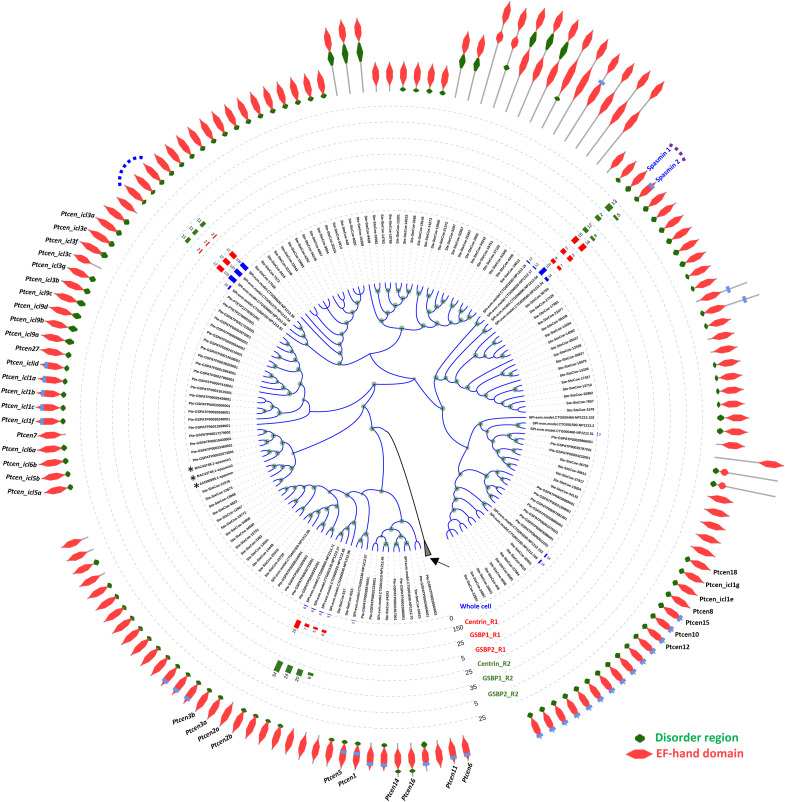
Fig. 2. Identification of spasmins in S. minus.
Phylogenetic tree showing all EF-hand proteins in S. minus, S. coeruleus, and P. tetraurelia. Asterisks indicate the three previously reported spasmins in Vorticella and Zoothamnium. Only EF-hand proteins closely clustered with spasmins in Vorticella and Zoothamnium are shown; the other EF-hand proteins (2087) are collapsed (black arrow). Predicted domain organization is assigned to each EF-hand protein. All the gene names of EF-hand proteins in Paramecium are labeled. Taxon names prefixed with “SPI,” “Ste,” and “Pte” are genes from S. minus, S. coeruleus, and P. tetraurelia, respectively. Bar chart shows the abundance (matched peptide number) of EF-hand proteins in MS or IP-MS experiments. Whole-cell MS analysis using S. minus cells. Centrin, IP-MS results using the anti-centrin antibody (mouse); GSBP1, IP-MS results using the anti-GSBP1 antibody (rabbit); GSBP2, IP-MS results using the anti-GSBP2 antibody (rabbit); R1 and R2, two replicates of IP-MS experiments. Dashed curve, EF-hand proteins found in the IP-MS results of GSBP1 and/or GSBP2. Two major EF-hand proteins, which show high abundance in whole-cell MS, centrin, GSBP1, and GSBP2 IP-MS, are named Spasmin 1 and 2, respectively.