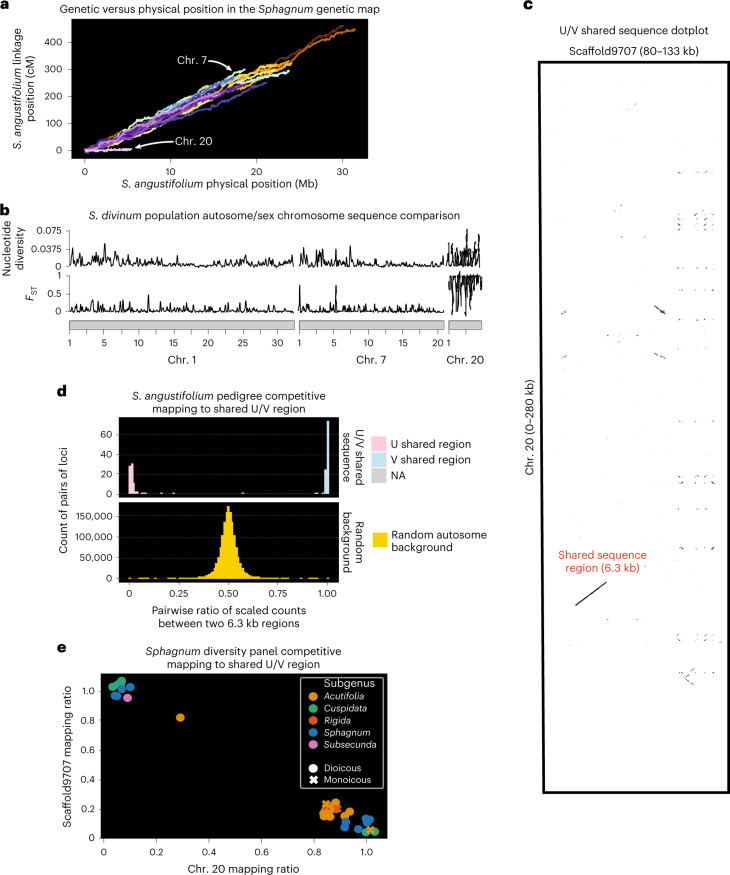
Fig. 4. U/V chromosome detection and analysis.
a, Recombination rate per chromosome, finding chr. 20 has a much lower rate of recombination than expected from the other 19 chromosomes. b, Sliding window analyses (100,000 bp window, 10,000 bp jump) of nucleotide diversity and FST between S. divinum chr. 20 SNP clusters. c, Exact k-mer dotplot with word size 15 for the shared sequence region between chr. 20 (putative V) and Scaffold9707 (putative U fragment), assembled from suspected female genotypes. d, S. angustifolium competitive mapping assay between chr. 20 and Scaffold9707. Ratio of reads mapped to the shared U/V region are shown, with individuals mapping to one sequence or the other (NA-ambiguous mapping ratio). Null distribution of autosome pairwise ratios is shown in yellow. e, Sphagnum diversity panel competitive mapping assay. Regardless of subgenera, individuals either mapped preferentially to the shared region of chr. 20 or Scaffold9707. Monoicous species (S. squarrosum, S. compactum, S. strictum and S. fimbriatum) each preferentially mapped to chr. 20. Positions on plot have been randomly ‘jittered’ by 0.1 units to improve readability among points.