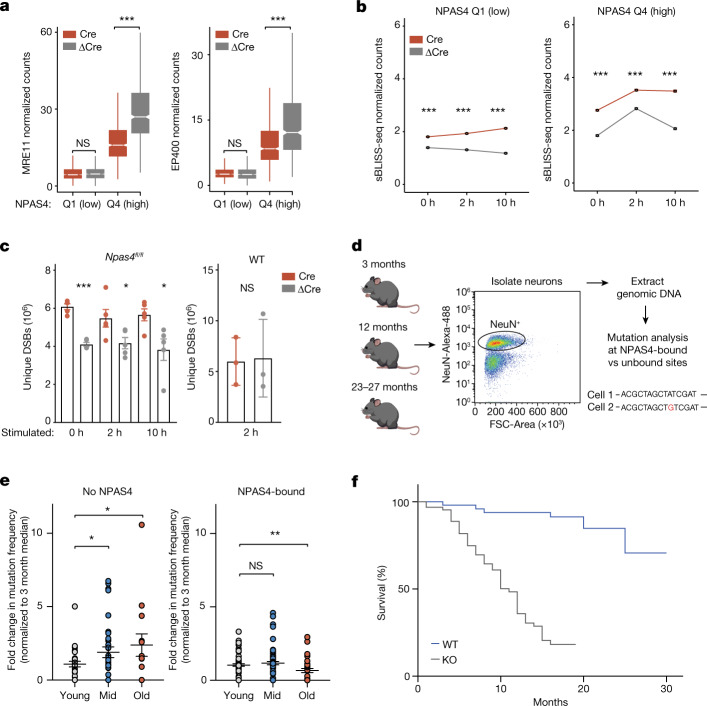
Fig. 5. NPAS4–NuA4 disruption impairs genome stability and reduces lifespan.
a, Boxplots of average MRE11 or EP400 CUT&RUN normalized counts in Cre-infected or ΔCre-infected hippocampi of Npas4fl/fl mice at activity-inducible sites, subset by quartiles of NPAS4 binding. Q1 = 1,017 sites, Q4 = 764 sites. Boxplots show the median (line), IQR (box) and 1.5× IQR (whiskers), and notches indicate the median ± 1.58× IQR/sqrt(n). n = 2–3 mice pooled per replicate. Replicate numbers provided in Supplementary Table 2. MRE11 Q1, P = 0.2453; Q4, ***P < 2.2 × 10−16. EP400 Q1, P = 0.9962; Q4, ***P < 2.2 × 10−16. P values were calculated using unpaired, one-tailed Wilcoxon rank-sum tests (ΔCre > Cre). b, Line plot depicting average ± s.e.m. of sBLISS-seq normalized counts in Cre-infected or ΔCre-infected hippocampi of Npas4fl/fl mice, subset by quartiles of NPAS4 binding. Q1 = 1,017 sites, Q4 = 764 sites. Data from n = 5 each for Cre-infected and ΔCre-infected mice. ***P < 2.2 × 10–16. P values were calculated using unpaired, one-tailed Wilcoxon rank-sum tests (Cre > ΔCre). See Extended Data Fig. 12a for boxplot distribution of data. c, Genome-wide breaks in hippocampal nuclei isolated from Npas4fl/fl (n = 5) or wild-type (n = 3) mice infected with Cre or ΔCre virus. Data are mean ± s.e.m. Npas4fl/fl: 0 h, P = 5.24 × 10–6; 2 h, P = 0.044; 10 h, P = 0.022. Wild-type: 2 h, P = 0.906. P values were calculated using two-tailed, unpaired t-tests. d, Collection of hippocampal neuronal nuclei across lifespan. e,Normalized mutation frequency at NPAS4-bound and unbound sites in wild-type mice. Mutation frequency (base changes and insertions and deletions) with age was calculated per site and normalized to the median frequency for that site in young animals. Points represent the normalized rate for one site sampled from one mouse. Data are mean ± s.e.m. Data are from n = 4 (young), 4 (middle aged) and 3 (old) mice. No NPAS4 sites: mid–young, *P = 0.03; old–young, *P = 0.016. NPAS4-bound sites: mid–young, NS = 0.18; old–young, **P = 0.0095. P values calculated using one-tailed, unpaired t-tests. f, Survival of Npas4 wild-type (n = 53; 28 females, 25 males) and Npas4 knockout (n = 64; 37 females, 27 males) mice, showing abbreviated lifespan of Npas4 knockout mice. P = 4.09 × 10–13 by a two-tailed Mantel–Cox test, P = 3.63 × 10–11 by a two-tailed Gehan–Breslow–Wilcoxon test.