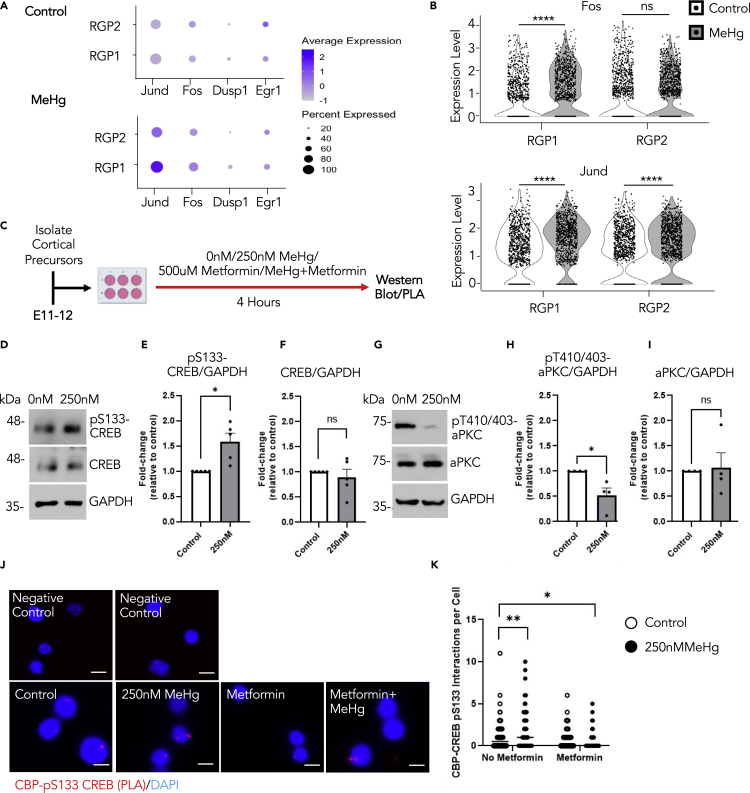
Figure 6.
Prenatal MeHg exposure stimulates CREB activity specifically in RGPs
(A) Dot plot of downstream CREB target genes (Jund, Fos, Dusp1, Egr1) and their expression from scRNA-seq data in RGP1 and RGP2 cell populations for control (0ppm) and 0.2ppm MeHg groups.
(B) Violin plot from scRNA-seq showing Fos (upper panels) and Jund (lower panels) expressions in RGP1 and RGP2 cell populations in control (0ppm) and 0.2ppm MeHg treatment groups, n > 800 cells for each group, Mann–Whitney U test, ∗∗∗∗p < 0.0001, ns (non-significant).
(C) Schematic of cortical percussors isolated from E11-12 CD1 mice, created with BioRender.com. Cells were exposed to 0 nM (control) or 250 nM MeHg with no metformin or metformin (500 μM) for 4 h, at which point they were lysed for western blot or PLA analysis.
(D–I) Western blot analysis of E12 cortical precursors cultured for 4 hours in the absence or presence of 250 nM MeHg. (D, G) Blots images were probed for pS133-CREB (D), pT410/403-aPKC (G), total CREB (D), total aPKC (G) and GAPDH (D, G) as a loading control. (E-F) Quantitative analysis of pS133-CREB (E) and total CREB (F) over GAPDH. n = 5 independent experiments, Student’s t test, ∗p < 0.05, ns (non-significant) (H-I) Quantitative analysis of pT410/403-aPKC(H) and aPKC (I) over GAPDH, n = 4 independent experiments, Student’s t test, ∗p < 0.05, ns (non-significant).
(J) Images of PLA+ interactions (lower panels, red) between pS133-CREB and CBP, counterstained with Hoechst (blue) in cultured cortical precursors of four treatment groups i) control (0ppm MeHg and 0 μM metformin), ii) 250 nM MeHg, iii) 500 μM metformin, and iv) co-treatment of 500 μM metformin +250 nM MeHg, scale bar: 5 μm. Images of negative control PLA (upper panels) in the absence of anti-pS133-CREB and anti-CBP (upper left) or absence of PLA probes (upper right).
(K) Quantitative analysis of pS133- CREB and CBP interaction signals (red) within nuclei (blue). n = 100 cells/group, n = 4 independent experiments, two-way ANOVA (group × metformin interaction F(1, 396) = 11.34, p = 0.0008, group F(1, 396) = 1.463, p = 0.2272, metformin F(1, 396) = 25.01, p < 0.0001, n = 400) with post hoc test, ∗p < 0.05, ∗∗p < 0.01. Error bars indicate the standard error of the mean (SEM). See also Figure S6.