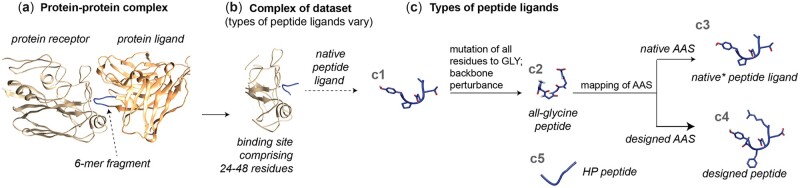
Fig. 1.
Generation of datasets of peptide–binding site complexes and overview of structures utilized in the study. (a) Crystal structure of the protein–protein complex with a selected interacting 6-residue fragment of protein–ligand. (b) Peptide–binding site complex of datasets. (c) Types of peptide ligands and their generation: native peptide ligand, the 6-residue fragment depicted in subfigure a (c1), was mutated into an all-glycine peptide ligand and perturbed (c2, Supplementary Information S1.3). Perturbed backbones were either reverted to their native amino acid sequence (c3, annotated as native* within the main text to highlight the backbone perturbation) or designed with PepSeP1 (c4). HP (highly perturbed) backbones (c5) are generated using the iNNterfaceDesign method (Syrlybaeva and Strauch, 2022)