Fig. 89.
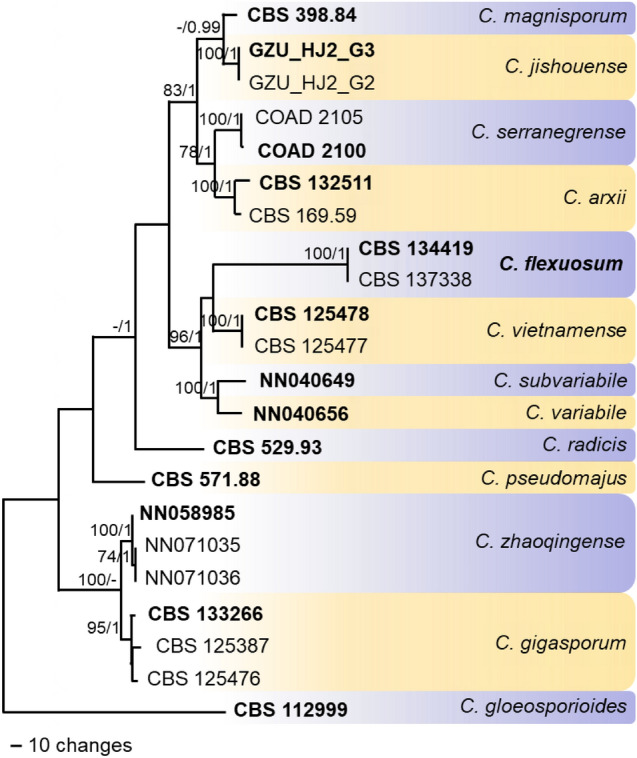
One of two most parsimonious trees obtained with PAUP v. 4.0b10 (Swofford 2003) from a heuristic search of the combined sequence alignment (gene boundaries of ITS: 1–550, tub2: 551–1240, gapdh: 1241–1510, act: 1511–1756, chs-1: 1757–2007, his3: 2008–2385) of the Colletotrichum gigasporum species complex, rooted with C. gloeosporioides CBS 112999 (sequences from Damm et al. 2012; Rakotoniriana et al. 2013; Liu et al. 2014; Silva et al. 2018; Zhou et al. 2019; Liu et al. 2022). Bootstrap support values (BS) above 70% (bold) and Bayesian posterior probability (BYPP) values above 0.90 are shown at the nodes. Bootstrap support values have been calculated based on 10 000 replicates, and a Markov Chain Monte Carlo algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v. 3.2.6 (Ronquist et al. 2012). Numbers of ex-type strains are in bold
