Fig. 2.
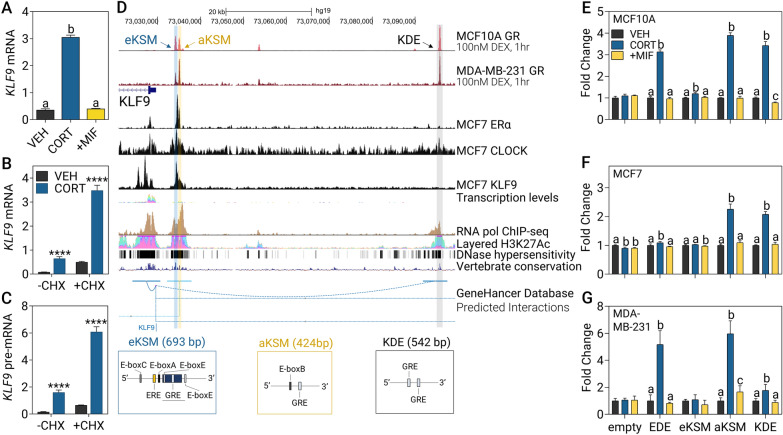
CORT-dependent induction of KLF9 is mediated by the GR through coordinate enhancer activation. A Pre-treatment with 1 μM of the GR-specific antagonist MIF for 1 h before addition of 100 nM CORT for 2 h abolished CORT-dependent KLF9 induction (one-way ANOVA; P < 0.0001). B, C MCF10A cells were pre-incubated with 100 μg/mL CHX for 30 min before treatment with 300 nM CORT for 2 h and KLF9 B mRNA and C pre-mRNA expression was measured through RT-qPCR (Student’s t-test; P < 0.0001). D The KLF9 locus and surrounding nongenomic regions were visualized in the UCSC genome browser [50] mapped to the GRCh37/hg19 assembly. Highlighted are eKSM, aKSM, and KDE, all located upstream of the KLF9 transcription start site. E–G The aKSM, eKSM, and KDE were cloned into the pGL4.23 luciferase construct and transfected into E MCF10A, F MCF7, and G MDA-MB-231 cells. An empty pGL4.23 vector were used as a negative control while the EDE [33] served as a positive control. Cells were then treated with vehicle (VEH), CORT (300 nM: MCF10A, MDA-MB-231; 1 μM: MCF7) or CORT plus MIF (1 μM; + MIF) for 20 h to evaluate GR-specific induction of luciferase activity for each enhancer (one-way ANOVA within a construct, followed by Tukey’s post-hoc test: P < 0.05; means with the same letter are not significantly different). Bars represent mean ± SEM and all experiments were performed with N ≥ 3 replicates