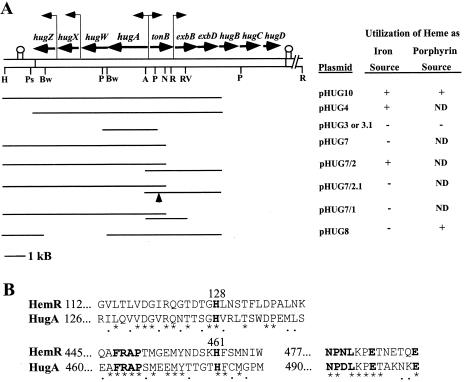
FIG. 1.
(A) Genetic and restriction enzyme map of cloned DNA from P. shigelloides heme iron utilization locus and identification of sequences required for utilization of heme as an iron source or as a porphyrin source. Genes carried by pHUG10 and pHUG16 are shown with horizontal arrows indicating the direction of transcription. Putative promoters are indicated with vertical bent arrows. Downstream of hugZ and hugD are regions of dyad symmetry indicated with two vertical lines with a circle at the top. The restriction enzyme sites shown are as follows: H, HindIII; Ps, PshA1; Bw, BsiWI; P, PstI; A, AseI; N, NheI; R, EcoRI; RV, EcoRV. Not all of the PstI, EcoRI, EcoRV, and PshAI sites are shown. The heme iron and heme porphyrin assays are described in Materials and Methods. ND, not determined. (B) Amino acid comparison of highly conserved heme receptor sequences in Y. enterocolitica HemR and P. shigelloides HugA. The numbers to the left of the sequences indicate the positions of the first amino acids. An asterisk below the sequence indicates identical amino acid residues and a period indicates conservative substitutions. The bold-faced sequences are those found by Bracken et al. (3) to be present in most heme receptors. Histidines 128 and 461 in HemR are important for the proper function of the protein (3).