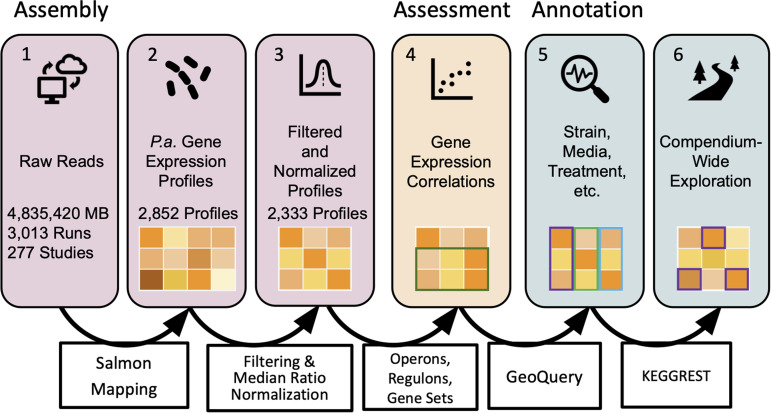
FIG 1.
Overview of the steps involved in compendium construction. The steps (numbered boxes) and corresponding methods (boxes with arrows) for assembly (steps 1 to 3), assessment (step 4), and annotation (steps 5 and 6) of a compendium of public RNA-seq gene expression profiles (represented by a matrix of orange squares) for P. aeruginosa are shown. (Step 1) Raw reads from the Sequence Read Archive (SRA) totaled over 4 million MB. “Runs” refers to fastq files, and “Studies” refers to the sets of runs deposited together. (Step 2) P. aeruginosa (P.a.) gene expression profiles refer to the results of mapping reads from a sample (referred to in the SRA as “experiment”) to a reference genome and can be read as counts or transcripts per million. (Step 3) Profiles were filtered to remove those that did not meet expression profile criteria and then median ratio normalized. (Step 4) Sets of coregulated genes were used to benchmark target patterns. (Step 5) Samples in the compendium were annotated for strain, media, genetic modifications, treatments, and other fields of interest. (Step 6) Pathway and function data facilitated compendium-wide explorations.