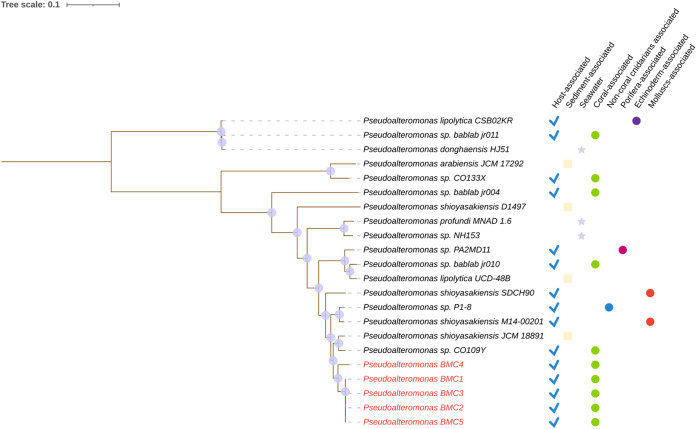
FIG 3.
Phylogenomic inference of publicly available genomes from 17 Pseudoalteromonas strains and the genomes of BMC strains 1 to 5 (in red), totaling 22 genomes. The tree was assembled from the comparison of 1,000 proteins through the codon tree method of the PATRIC platform that selects global protein families (PGFams) as homology groups and analyzes aligned proteins and DNA encoding single-copy genes using the RAxML program. The best protein model found by RAxML to build this tree was Jones-Taylor-Thornton - Direct Computation with Mutabilities (JTTDCMut). Purple dots on the branch length correspond to the bootstrap value of 100. The icons on the right side of the strains represent their isolation sources.