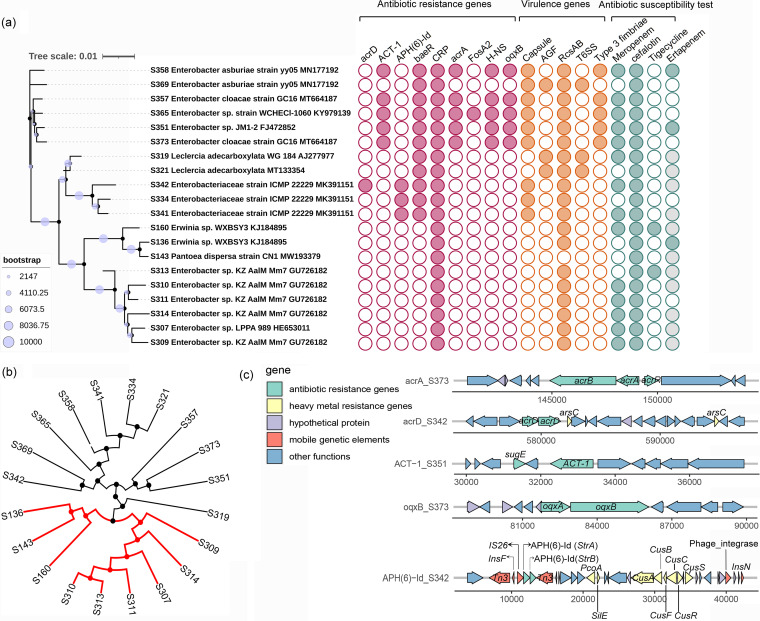
FIG 5.
Genomic analysis and antimicrobial susceptibility of Enterobacteriaceae species (n = 20) retrieved from floors and escalators. (a) Phylogenetic tree (left) of all strains based on full-length 16S rRNA gene sequences (bootstrap = 10,000). A bubble plot (right) reflects the detection of antibiotic resistance genes (annotated by searching against CARD database; red) and virulence genes (annotated by searching against VFDB database; orange) from genome sequences and antimicrobial susceptibility test (green; strains marked in gray are the ones for which the ertapenem susceptibility test was not performed). (b) A rooted maximum-likelihood phylogenetic tree was constructed based on the alignment of the core genome single nucleotide polymorphisms. The strains in the red branches clustered together, which was consistent with the clustering of their full-length 16S rRNA genes. (c) Flanking regions of the detected antibiotic resistance genes in contigs. Arrows indicate the direction of gene transcription. Different genes are indicated by different colors. Genes with ≥98% amino acid identity and a query coverage of ≥99% were annotated by mapping sequences against the NCBI nr database. AGF, curli fibers/thin aggregative fimbriae.