Figure 9.
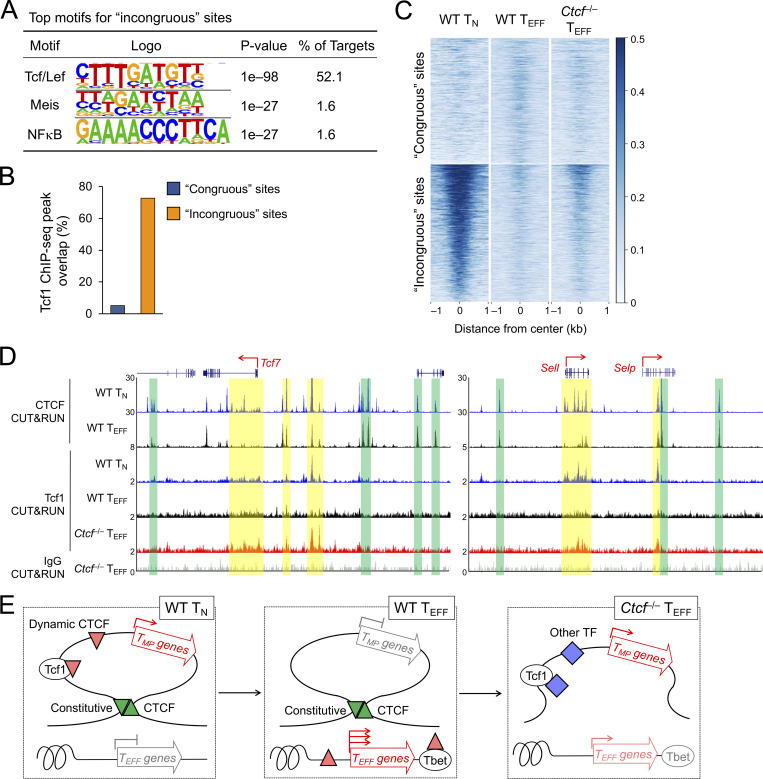
Constitutive CTCF binding limits access to TMP genes by Tcf1. (A) Top motifs of the incongruous CTCF sites (defined in quadrant iii in Fig. 6 C), with the congruous sites as background based on HOMER analysis. (B) Distribution of Tcf1 ChIP-seq peaks in the congruous and incongruous sites. (C) Heatmaps show Tcf1 binding strength at the congruous (top) and incongruous (bottom) sites, as detected with CUT&RUN in WT TN, WT and Ctcf−/− TEFF cells. (D) Tracks of CTCF and Tcf1 CUT&RUN in TN or TEFF cells at the Tcf7 and Sell gene loci. The CTCF binding tracks in WT TN and TEFF cells are the same as Fig. 8 E as reference for the boundary anchors (denoted with green bars). Yellow bars highlight increased Tcf1 binding signals in Ctcf−/− over WT TEFF cells. Note that Tcf1 binding in Ctcf−/− TEFF cells remained weaker than that in WT TN cells, as displayed on different y-axis scales. (E) Diagram showing the proposed model on the cooperative actions of constitutive and dynamic CTCF binding in promoting TEFF cell differentiation. Genes in the TEFF program acquire novel CTCF binding and increased ChrAcc (upward red triangles), directly or indirectly by Tbet or other TFs (such as those in AP1, Runx, and Ets families, not depicted). On the other hand, genes in the TMP program are positioned in insulated neighborhoods, represented by a single loop demarcated with convergent, constitutive CTCF binding sites (green triangles). The structure of the insulated neighborhoods is preserved in TN and TEFF cells, and the downregulation of TMP genes is likely controlled locally, associated with Tcf7 gene silencing and eviction of CTCF binding (downward red triangle) within the insulated neighborhoods. Genetic ablation of CTCF impairs dynamic CTCF-mediated activation of TEFF genes and disrupts constitutive CTCF-organized insulated chromatin structure, resulting in access to TMP genes by activating TFs (blue diamonds) including Tcf1.