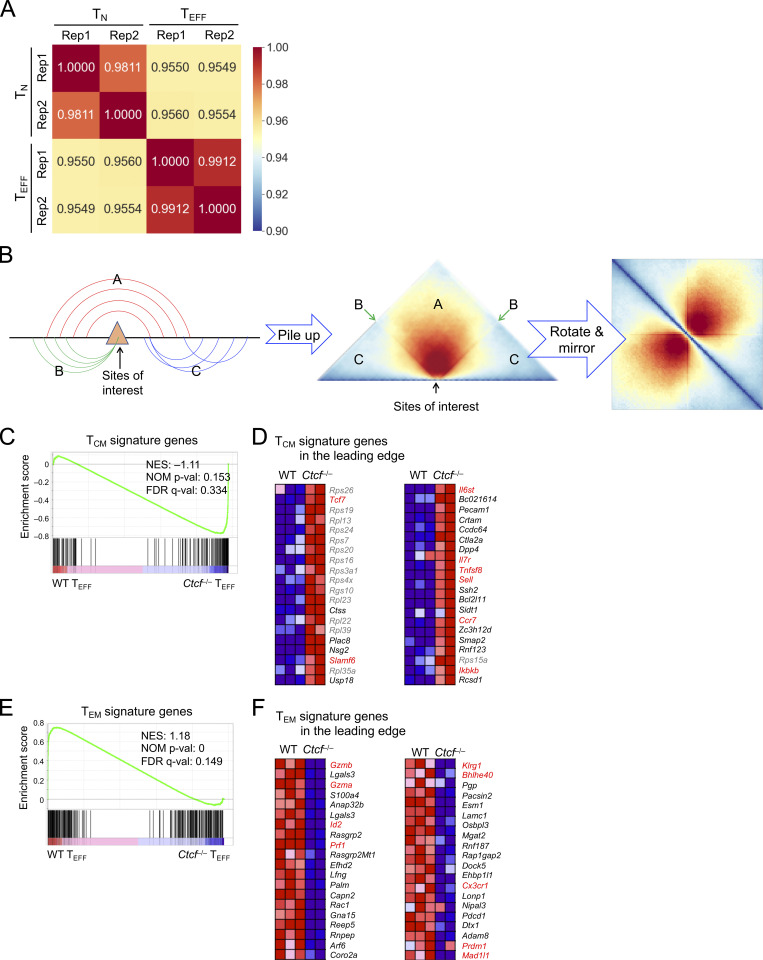
Figure S3.
Hi-C data characterization and GSEA of TCM and TEM signature genes. (A) Correlation heatmap showing reproducibility of Hi-C libraries from TN and TEFF cells. Pearson correlation coefficient between Hi-C library pairs was determined using ChrInt scores at 10 kb resolution as the input data (see details in Materials and methods). (B) Diagram illustrating the summation approach underlying the Hi-C pile-up profile centered on specific genomic locations (sites of interest). The ChrInt between the regions flanking the sites of interest is aggregated in section A, while that within the region on either side of the sites is in sections C. The interaction of the sites with its flanking regions is projected on lines B. (C–F) RNA-seq data on TCM and TEM cells were retrieved and processed in-house. By requiring ≥1.5-fold changes, FDR < 0.05, and fragments per kilobase of exon per million mapped fragments at the higher expression condition ≥1, we identified 144 genes as TCM signature and 231 genes as TEM signature genes. The enrichment of these signature genes in WT and Ctcf−/− early TEFF transcriptomes was determined with GSEA. (C and D) TCM signature is enriched in Ctcf−/− early TEFF cells, with 49 TCM signature genes in the leading edge. C, enrichment plot; D, heatmaps of the top 40 genes in the leading edge, with genes of interest in red font. (E and F) TEM signature is enriched in WT early TEFF cells, with 44 TEM signature genes in the leading edge. E, enrichment plot; F, heatmaps of the top 40 genes in the leading edge, with genes of interest in red font. Marked in enrichment plot are NES (normalized enrichment score), NOM P val (nominal P values), and FDR q-val (FDR q values) as output from GSEA. Note that similar results were obtained with TCM and TEM signature gene sets defined in GSE147080 (Millner et al., 2020).