Abstract
The field of 3D genome organization produces large amounts of sequencing data from Hi-C and a rapidly-expanding set of other chromosome conformation protocols (3C+). Massive and heterogeneous 3C+ data require high-performance and flexible processing of sequenced reads into contact pairs. To meet these challenges, we present pairtools – a flexible suite of tools for contact extraction from sequencing data. Pairtools provides modular command-line interface (CLI) tools that can be flexibly chained into data processing pipelines. Pairtools provides both crucial core tools as well as auxiliary tools for building feature-rich 3C+ pipelines, including contact pair manipulation, filtration, and quality control. Benchmarking pairtools against popular 3C+ data pipelines shows advantages of pairtools for high-performance and flexible 3C+ analysis. Finally, pairtools provides protocol-specific tools for multi-way contacts, haplotype-resolved contacts, and single-cell Hi-C. The combination of CLI tools and tight integration with Python data analysis libraries makes pairtools a versatile foundation for a broad range of 3C+ pipelines.
Intro
Chromosome conformation capture technologies (3C+), particularly Hi-C, revolutionized the study of genome folding by using high-throughput sequencing to measure spatial proximity. In all 3C+ approaches, spatial proximity is captured via DNA ligation, resulting in libraries of chimeric DNA molecules. Contact frequency between pairs of genomic loci is then read out by sequencing these libraries. All 3C+ protocols involve four key steps: (i) chemical cross-linking of chromatin 1, (ii) partial digestion of DNA followed by (iii) DNA ligation, and (iv) sequencing 2. Ligation is the pivotal step that records the spatial proximity of DNA loci in chimeric DNA sequences. In a typical 3C-based experiment, the DNA ligation products are subsequently shredded into smaller pieces, purified, and amplified. The resulting libraries are typically sequenced in short-read paired-end mode (around 50–300 bp on each side), producing millions to tens of billions of sequencing reads.
Processing 3C+ sequencing data requires highly-performant and specialized computational tools. First, the quantity of 3C+ data is increasing rapidly. A growing number of labs and consortia (4DN 3, ENCODE 4, DANIO-CODE DCC 5) use Hi-C to produce large quantities of proximity ligation data. At the same time, new protocols, such as Micro-C 6 and Hi-C 3.0 7, improve resolution and sensitivity and generate even larger datasets. This requires software to be fast, parallelizable, and storage-efficient. Emerging 3C+ technologies introduce additional challenges, including measuring contacts within individual homologs 8 and sister chromatids 9 10, single cells 11 12 13 14 15, and multi-way contacts (Pore-C16 and MC-3C 17). This large variation in the growing family of 3C+ methods thus requires software to be versatile and flexible.
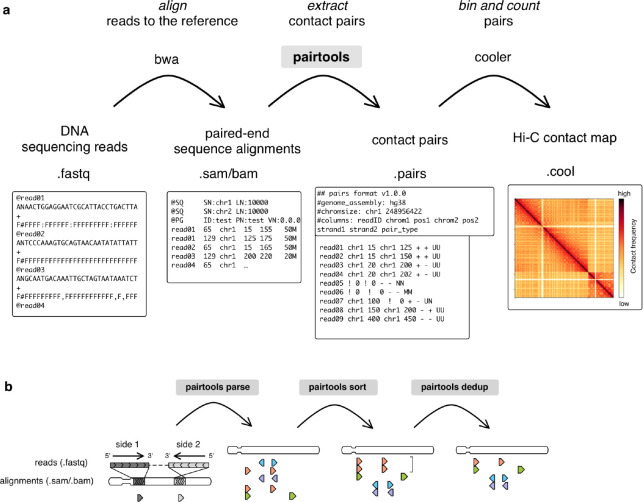
3C+ data are typically computationally processed in three stages (Figure 1a). First, sequencing reads are aligned against the reference genome. Next, pairs of genomic locations are extracted from the alignments. These pairs may be interpreted as genomic contact events. For various statistical and technical reasons, pairs are normally aggregated or binned to form contact matrices at various lower genomic resolutions. Binned contact maps can be stored, manipulated, and analyzed using downstream tools and software packages, such as cooler 18 and cooltools 19 from Open2C. However, a great deal of pertinent experimental information in 3C+ data accessible at the level of pairs is lost during the process of binning. This includes classification and quality control information, such as exact mapping positions, strand orientation, mapping quality, and pair type. As a result, pairs serve as a resource for generating custom filtered contact matrices. For these reasons, it is important to be able to flexibly generate, interpret, store, and manipulate pairs-level data.
Figure 1. Processing 3C+ data using pairtools.
a. Outline of 3C+ data processing leveraging pairtools. First, a sequenced DNA library is mapped to the reference genome with sequence alignment software, typically using bwa mem for local alignment. Next, pairtools extracts contacts from the alignments in .sam/.bam format. Pairtools outputs a tab-separated .pairs file that records each contact with additional information about alignments. A .pairs file can be saved as a binned contact matrix with counts with other software, such as cooler. The top row describes the steps of the procedure; the middle row describes the software and chain of files; the bottom row depicts an example of each file type.
b. Three main steps of contact extraction by pairtools: parse, sort, and dedup. Parse takes alignments of reads as input and extracts the pairs of contacts. In the illustration, alignments are represented as triangles pointing in the direction of read mapping to the reference genome; each row is a pair extracted from one read. The color represents the genomic position of the alignment with the smallest coordinate, from the leftmost coordinate on the chromosome (orange) to the rightmost coordinate on the chromosome (violet). Sort orders mapped pairs by their position in the reference genome. Before sorting, pairs are ordered by the reads from which they were extracted. After sorting, pairs are ordered by chromosome and genomic coordinate. Dedup removes duplicates (pairs with the same or very close positions of mapping). The bracket represents two orange pairs with very close positions of mapping that are deduplicated by dedup.
Here we introduce Pairtools, a suite of flexible and performant tools for converting sequenced 3C+ libraries into captured chromosome contacts. Pairtools provides modules to (i) extract and classify pairs from sequences of chimeric DNA molecules, (ii) deduplicate, filter, and manipulate resulting contact lists, and (iii) generate summary statistics for quality control (QC). Pairtools enables the processing of standard Hi-C 2 as well as many Hi-C-derived protocols, including homolog- and sister-sensitive, multi-contact, single-cell Hi-C protocols. Pairtools uses a well-defined data format and may be arranged in highly efficient data pipes (Figure 1a, b). The benchmark against several popular 3C+ data mappers shows advantages of pairtools for high-performance and flexible 3C+ analysis. Pairtools is implemented in Python, powered by common data analysis libraries such as NumPy 20 and pandas 21, offers a CLI, and is available as open-source software at: https://github.com/open2c/pairtools/.
Design principles
Pairtools provides tools for each step of data processing between the sequence alignment and the contacts binning (Figure 1a, b): extraction, processing, filtering, and quality control of contact pairs.
Pairtools adheres to the following design principles, aligned with Unix style principles 22:
Split functionality into tools that can be used independently or combined into pipelines.
Focus on modularity, flexibility, and clarity first and performance second. Data processing should be as fast as alignment, but not necessarily faster.
Outsource functionality when possible. Rely on existing software for alignment and workflow managers for data pipelining.
Leverage the rich ecosystem of Python and data analysis libraries, including NumPy 20, pandas 23, scipy 24 and scikit-learn 25.
Use a standardized tabular format for pairs.
Accommodate existing Hi-C protocol modifications by generalizing existing tools. When not possible, introduce protocol-specific tools.
Take advantage of multi-processing and streaming to improve performance.
Essential building blocks for 3C+ pair processing
Pairtools processes 3C+ data in three essential steps (Fig 1b). First, the genomic alignments of 3C+ products are parsed into individual contact events, or pairs. Second, the resulting pairs are sorted to facilitate data access and analysis. Third, pairs are deduplicated, resulting in the final list of 3C+ contacts.
The minimal pairtools-based pipeline is expressed concisely as:
bwa mem index input.R1.fastq input.R2.fastq \
| pairtools parse -c chromsizes.txt \
| pairtools sort \
| pairtools dedup \
| cooler cload pairs -c1 2 -p1 3 -c2 4 -p2 5 chromsizes.txt:1000 - output.1000.cool
Below, we describe these three steps and the corresponding pairtools functionality in detail.
parse: extracting single proximity ligation events
The DNA molecules in a 3C+ library are chimeric by design: spatial proximity between different genomic segments is captured as DNA ligation events, which are then read out via DNA sequencing. Pairtools makes use of existing software for alignment, taking .sam/.bam files 26 as input. Sequence alignments in .sam/.bam comprehensively describe the structure of chimeric DNA molecules in the 3C+ library. Each entry in these files stores an alignment of a continuous read segment to the reference genome. Entries include mapping position, as well as flags and tags describing mapping properties (such as uniqueness of mapping, nucleotide variations, error probability, and more). The properties can be read with tools like pysam 27. However, alignments in the .sam/.bam files are reported sequentially and are not structured as contact pairs. Extraction of proximity ligation events from alignments requires additional processing, which pairtools achieves with parse.
Pairtools parse is developed and optimized for Hi-C libraries with chimeric DNA molecules formed via single ligation events. Pairtools relies on local sequence aligners (e.g., bwa mem) that can split chimeric reads into segments, each aligned to a different location in the genome. Pairtools parse extracts pairs of adjacent alignments and reports them as individual contacts, categorizing them based on uniqueness (Fig 2a, Supplementary Figure 1a–c) and exposing the properties of alignments relevant to Hi-C. Parse also detects cases when one of the DNA fragments in a pair was sequenced on both sides of the read and thus is not a contact but an artifact of paired-end sequencing. Parse detects such cases, and merges the alignments, “rescuing” the true contact pair (Supplementary Figure 1d). The output of parse adheres to the standard format .pairs 28, discussed below.
Figure 2. Parsing contact pairs and walks from alignments.
a. Parsing is the first step of contact extraction and can be done by either parse or parse2 in pairtools. The choice of the tool is directed by the read length and the abundance of multiple ligation events. For single ligation events, pairtools reports the type of pair (in the example here, both alignments are unique, UU, and other types listed in Supplementary Figure 1a–g). For multiple ligation events, pairtools distinguishes the ligation type of the pair (walk_pair_type: R1 and R2 - direct ligations observed on the first or the second side of the read; R1–2 unobserved ligation that can be potentially indirect), and reports the order of pair in a walk (walk_pair_index). For other examples, see Supplementary Figure 1d–g.
b. Resolving multi-way contacts with 3C+ methods. In a 3C+ library, a multi-way contact is captured as a chimeric DNA molecule. Each end of DNA fragment can be ligated to its neighbor only once, i.e. hop to another DNA fragment. Contacts between consecutively ligated fragments of the chimera are called “direct” (i.e., directly ligated, 1-hops); those between non-adjacent fragments are “indirect” (2-, 3- and many-hops). In paired-end sequencing, a fraction of the molecule remains unsequenced and may contain DNA fragments (missing contacts). As a result, the contacts between the two fragments abutting the gap are called “unobserved”, and they may be either direct or indirect.
c. Contacts recovery relative to default pairtools settings for different parsing modes of long walks in paired-end 3C+ data with reads ~150–250 bps (data from 9, 38, 39, 40).
The engine of pairtools parse uses pysam 27 to extract tags and flags from .sam/.bam files. Pairtools parse can run in combination with a variety of popular local sequence aligners, such as bwa mem 29, bwa mem2 30, minimap2 31, and others, as long as their output complies with the .sam/.bam format.
parse2: extracting multiple proximity ligation events
Early Hi-C protocols aimed to produce libraries of chimeric molecules containing single ligation sites. However, newer experiments routinely produce molecules containing multiple ligation events, also known as multi-way contacts (Figure 2b, Supplementary Figure 1d–g), for the following reasons:
The latest 3C+ protocols use more frequently cutting restriction enzymes (and their combinations 7) or non-specific endonucleases 32, resulting in smaller ligated DNA fragments 1 and more reads from multiple ligation events.
The technologies for paired-end “short read” Illumina sequencing can now generate reads of several hundred nucleotides per side, which often exceeds the average fragment length.
More dramatically, new 3C+ protocols use long-read sequencing technology to purposefully generate molecules with multiple ligation sites to detect co-occurring proximity events 16 17 33 34.
The increasing prevalence of datasets with multi-way contacts 35 36 calls for a tool dedicated to their extraction. This process differs substantially from the extraction of single proximity ligation events.
First, the structure of multi-way contact is much more complex than a single proximity event (Figure 2b). Each library DNA molecule is expected to contain multiple fragments connected by pairwise direct ligation junctions. Multi-way contacts are ordered by the position within the chimeric DNA molecule. Since the order of the ligation events along a 3C+ chimeric molecule can contain meaningful information, multi-way contacts with a defined order are referred to as walks 37 (Figure 2b). For example, this information is used to quantify either chromosomal entanglements or traversals between A and B compartments 17 with walks.
Further complexity arises when the contacts are categorized by the number of ligation events. When two fragments are sequenced one after another on the same read, they form direct ligation because the ligation junction between them was observed (Figure 2b). They are also called single hops because there is a single disruption of continuous DNA fragment 37. Some types of 3C+ analyses augment the data to include indirect ligations (mediated by multiple ligation events, two-, three- and many-hops) by combinatorial combinations of fragments in the same walk 16 36.
Long chimeric DNA molecules can be sequenced by either traditional paired-end or single-end long-read sequencing. Paired-end sequencing introduces additional complications and uncertainty into analysis.
In paired-end sequencing, if the 3C+ library DNA molecules are much longer than two read lengths, then the central side of the DNA molecule can remain unsequenced (Figure 2b, Supplementary Figure 1e–f). This insert can potentially contain other DNA fragments, resulting in indirect ligation. Thus, contacts formed by the ends of paired reads should be interpreted as unobserved ligations.
The opposite scenario in paired-end sequencing is when the length of the DNA molecule is smaller than two read lengths, resulting in a read-through. The same subregion of a molecule is sequenced twice, producing internal duplicates (Supplementary Figure 1g).
Pairtools parse2 extracts multi-way contacts from single 3C+ chimeric molecules resolving these issues (Figure 2a) and provides rich and flexible reporting (Supplementary Figure 1h). Parse2 works with both paired “short” reads and long read technologies and has the following features:
Parse2 preserves and reports the order of ligation events (a walk) within a molecule (Figure 2a).
Parse2 removes internal duplicates ensuring pairs are not reported twice if they were sequenced by the overlapping reads on both sides of the molecule (Supplementary Figure 1g).
Parse2 distinguishes direct and unconfirmed contacts and reports the type, e.g., originating from the first side of paired-end read (R1), the second side (R2), present on both sides (R1&R2), or formed by end alignments on two sides (R1-R2) (Figure 2a, Supplementary Figure 1e–g). Parse2 reports this information and, therefore, allows filtration of only direct ligation events.
Parse2 can combinatorially expand contacts of a walk by generating all pairs of DNA fragments of the same molecule, including indirect ones 16.
Parse2 provides flexibility for reporting orientation and mapping positions of alignments in pairs. Pairs can either be reported either traditionally (as 5’-ends of alignments) or as ligation junctions (precise nucleotide-resolved positions of contacting alignment ends) (Supplementary Figure 1b).
While parse now supports some of these features for backward compatibility reasons, we strongly recommend using parse2 to analyze multi-contact data, such as PORE-C 16 and MC-3C 17 (Figure 2a). When applied to regular Hi-C or Micro-C, parse2 captures more contacts than regular parse (Figure 2c) because it captures the contacts originating from walks. We observe a substantial gain (5–25%) of contacts for both 150 bps 3C+ reads 38 39 and 250 bps long reads 9. A common approach is to include all pairwise contacts within each walk via combinatorial expansion 16. However, we do not recommend this method because we find evidence of less desirable properties of indirect contacts generated this way (see section on quality control below).
Pairtools uses and extends the .pairs format
Both parse and parse2 output contact tables in a text tab-separated format called .pairs, designed by the NIH 4DN consortium 28. As a text format, .pairs has several advantages over custom binary formats: (i) text tables can be processed in all programming languages, (ii) are easily piped between individual CLI tools, and (iii) have a set of highly efficient utilities for sorting (Unix sort), compression (bgzip/lz4), and random access (tabix 41 /pairix 28).
Each tab-separated row in a .pairs file describes a single observed contact. The required columns contain the id of the read and the genomic locations of the two sites that formed the contact. Pairtools parse augments these data with the pair type (Supplemental Figure 1a–g) and optional columns with details of the genomic alignments supporting the contact.
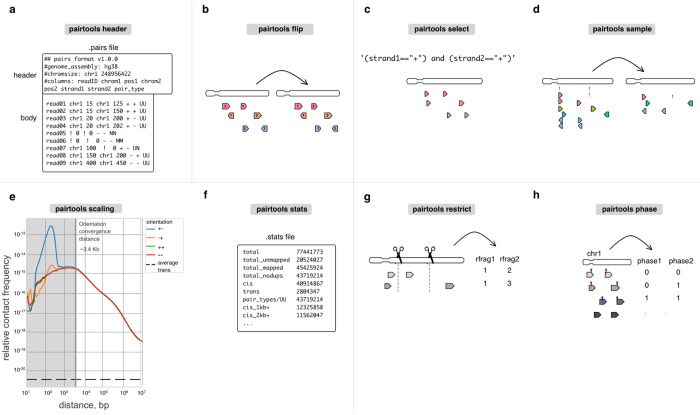
Headers of .pairs files can store metadata, which by default includes the names of columns and the description of chromosomes of the reference genome. To ensure data provenance, pairtools extends this standard header with (i) the header of the .sam files that stored the original alignments and (ii) the complete history of data processing, with a separate entry for each CLI command of pairtools that was applied to the file. Pairtools provides a set of CLI and Python functions to parse and manipulate the header. Pairtools header can generate a new header, validate an existing one, transfer it between .pairs files, as well as set new column names (Figure 3a).
Figure 3. Auxiliary tools for building feature-rich pipelines.
a. Header verifies and modifies the .pairs format.
b-d. Flip, select, and sample are for pairs manipulation.
e-f. Scaling and stats are used for quality control. For scaling, we report scalings for all pairs orientations (+−,−+, ++, −−) as well as average trans contact frequency. Orientation convergence distance is calculated as the last rightmost genomic separation that does not have similar values for scalings at different orientations.
g-h. Restrict and phase are protocol-specific tools that extend pairtools usage for multiple 3C+ variants.
sort and flip: organizing contact lists
Sorting the pairs in contact tables facilitates data processing, and analyses as it (i) enables fast random access via indexing and (ii) simplifies the detection of duplicated pairs, as they end up in adjacent rows of the table. Pairtools sorts pairs in two steps. First, individual pairs are “flipped,” i.e., arranged so that the alignment with the lower coordinate in the genome comes first in the pair (Figure 3b). Flipping is necessary as the two sides of a DNA molecule are sequenced in random order. Flipping is performed by default during parsing and can be done manually by pairtools flip. Second, pairtools sort performs Unix-based sorting of pairs in contact tables according to their genomic positions (on chrom1, chrom2, pos1, pos2). This sorting scheme has multiple advantages: it arranges pairs in blocks corresponding to contacts between a given pair of chromosomes, separates within (cis) from between (trans) chromosome contacts, and facilitates access to unmapped and multi-mapped pairs.
dedup: detecting duplicated DNA molecules
A key issue for sequencing-based protocols, including Hi-C and other 3C+, is that the same DNA product can be duplicated by PCR and then sequenced and reported more than once, thus introducing an error into their quantitative measurements. Pairtools provides a computationally efficient tool for duplicate removal called pairtools dedup (Figure 1b). It detects clusters of pairs with matching genomic coordinates and strands and removes all but one pair. Pairtools dedup additionally permits minor mismatches between coordinates to account for potential losses of a few nucleotides from the ends of a molecule. To enable such imperfect coordinate matching, pairtools dedup relies on a KD-tree-based fixed radius nearest neighbor search algorithm 42, implemented in scipy and scikit-learn. To reduce its memory footprint, pairtools dedup processes input data in overlapping chunks. Finally, pairtools dedup can take additional columns from a .pair file and require additional properties of pairs to match, such as type of contact (direct/indirect), presence of mutations, or phased haplotype 8 ).
Tracking duplicates with pairtools enables an estimate of library complexity, i.e. the total number of unique DNA molecules prior to PCR amplification, an important QC for 3C+. Library complexity can guide the choice of sequencing depth of the library and provide an estimate of library quality. To estimate library complexity, pairtools assumes that each sequencing read is randomly chosen with replacement from a finite pool of fragments in DNA library 43 44.
Tools for building feature-rich 3C+ pipelines
In addition to supporting the parse-sort-dedup steps (Figure 1b) that are sufficient to build a minimalistic 3C+ processing pipeline, pairtools also provides tools to build feature-rich pipelines. This includes tools for automated QC reporting, filtering of high-quality 3C+ data, as well as merging of replicates and conditions into meta contact maps, required for a complete and convenient end-to-end data processing pipeline.
select, sample, and merge: manipulating pairs
Pairtools provides a collection of tools for the manipulation of tabular pairs data.
pairtools select splits and subsets datasets according to arbitrary filter expressions (Figure 3c). These expressions are provided as Python functions, enabling expressive and powerful filters. Filters can include wildcard and regex matching on string columns, custom functions from 3rd-party libraries (for examples of advanced usage, see Sister-C 10, scsHi-C 9), as well as filtering pairs to a given subset of chromosomes.
pairtools sample can generate random subsets of pairs, e.g., to equalize coverage between libraries or assess the statistical robustness of analyses (Figure 3d).
pairtools merge combines multiple input datasets into one; for pre-sorted inputs, it efficiently produces sorted outputs.
Together, pairtools select and merge enable the split-apply-combine pattern for distributed data processing.
stats and scaling: quality control
3C+ experiments have multiple failure modes and thus require tight quality controls. Many experimental issues can be inferred from the statistics of the resulting 3C+ data (for a detailed discussion, see 45 46).
A particularly rich source of information about 3C+ experiments is the decay of contact frequency with the genomic distance referred to as the P(s) 2 curve or scaling (borrowing the physics terminology for power-law relationships). Scalings are used both to characterize mechanisms of genome folding 19 and reveal issues with QC 1. For example, early flattening of the scaling in Micro-C revealed the importance of long cross-linkers 6. Scalings can also be used to determine that combinatorial expansion of walks produces undesirable contacts because indirect contacts result in flatter scaling (Supplementary Figure 2e) 17 16.
Pairtools scaling calculates strand-oriented scalings that can be used for by-product quality control and filtration (Figure 3e). After the ligation step, some fragments can form a valid pair or produce unwanted 3C+ by-products, such as self-circles, dangling ends (unligated DNA) (Supplementary Figure 2c), and mirror reads (potential PCR artifacts) 47. A short-range peak in divergent orientation is a sign of self-circled DNA, while a short-range peak in convergent orientation is a sign of dangling ends (Figure 3e, Supplementary Figure 2d) 45 46. For example, early Hi-C variants with a low concentration of cross-linker caused the prevalence of self-circles 48. At larger genomic separations, pairs are formed in all four orientations with equal probabilities, and strand-oriented scalings converge. The orientation convergence distance indicates the minimum distance where pairs can simply be interpreted as contacts for a 3C+ dataset. Removing contacts below the orientation convergence distance removes nearly all by-products marked by restriction fragment annotation (see below, Supplementary Figure 2b). For DpnII Hi-C datasets, orientation convergence usually occurs by ~2kb. We note that for analyses downstream of QC, scaling can also be calculated from corrected binned data, e.g., using cooltools 19.
For convenience and workflow reproducibility, pairtools stats automatically reports contact scalings for individual chromosomes. It also generates additional summary statistics, including the total number of pairs based on their type, and the number of trans contacts. This has been used to understand the impact of various protocol decisions. For example, information about the frequency of trans and different ranges of cis-contacts demonstrated that extra cross-linking yields more intra-chromosomal contacts 1. The frequency of contacts between the nuclear and mitochondrial genomes reflects the noise introduced by various digestion strategies 1. pairtools stats produces a human-readable nested dictionary of statistics stored in a YAML file or a tab-separated text table (used in 49, 50) (Figure 3f). These outputs can be visualized with MultiQC 51, an interactive web-based tool that aggregates a wide set of sequencing QC statistics and provides an overview of whole collections of samples.
Protocol-specific tools
Chromosome capture is a growing field, with novel protocol modifications emerging regularly 52. Thanks to its flexible architecture, pairtools can process data from many such experiments. For example, data from chemical modification-based protocols, such as scsHi-C 9, sn-m3C-seq 53, or Methyl-HiC 54 can be processed by (i) extracting sequence mismatches into separate columns of .pairs by pairtools parse and (ii) filtering and subsetting pairs based on these columns with pairtools select (Figure 3c). For other popular and actively developing protocol variants, such as Micro-C 6, haplotype-resolved 8 and single-cell Hi-C 55 , pairtools provides specialized utilities.
restrict: annotating pairs by restriction fragments
Many 3C+ protocols, particularly original Hi-C, rely on cutting DNA by restriction enzymes and theoretically should generate ligations only between restriction sites 45 56. Thus, early 3C+ analysis pipelines included filters that detected and removed (i) unligated single restriction fragments and (ii) ligations between pieces of DNA located far from any restriction sites. Pairtools restrict enables such filters by assigning the nearest restriction sites to each alignment in a pair (Figure 3g).
However, we find restriction-based filters unnecessary for more recently-published Hi-C and do not include them in the standard pairtools pipeline. First, in our tests of recently published Hi-C datasets 1, the statistical properties of pairs located far from and close to restriction sites proved nearly the same (Supplementary Figure 2a). Second, we found that unligated pieces of DNA can be removed by a simpler filter against short-distance pairs, which can be calibrated using strand-oriented scalings 46 (Supplementary Figure 2b).
For downstream analyses in cooler 18, such by-products are removed by dropping pairs of bins with separations below a cutoff distance, which corresponds to removing a few central diagonals of a binned contact matrixf. Finally, the annotation of restriction sites becomes less accurate and unproductive for libraries generated with frequent and/or flexible cutters (e.g., DpnII, MboI, and DdeI), cocktails thereof, and impossible in restriction-free protocols, such as Micro-C 6 and DNase Hi-C 57.
phase: annotating haplotypes
Haplotype-resolved Hi-C 8 58 59 60 61 leverages sequence variation between homologous chromosomes to resolve their contacts in cis and trans. In particular, single nucleotide variants (SNVs) can be used to differentiate contacts on the same chromosome (cis-homologous) from contacts between two homologs (trans-homologous).
Pairtools phase is designed to resolve reads mapped to maternal and/or paternal genomes (Figure 3h). To enable analyses with pairtools phase, reads must be mapped to a reference genome that contains both haplotypes, reporting suboptimal alignments; these suboptimal alignments will be extracted into separate .pairs columns by pairtools parse. By checking the scores of the two best suboptimal alignments, pairtools phase distinguishes true multi-mappers from unresolved pairs (i.e., cases without a distinguishing SNV/indel) and reports the phasing status of each alignment in a pair: non-resolved, resolved as the first haplotype or the second haplotype, or multi-mapper. Further downstream, pairtools select and pairtools stats enable splitting and analyzing phased pairs. Pairtools phase has been successfully used to study allele-specific folding of chromosomes in D. melanogaster and mouse 8.
filterbycov: cleaning up single-cell data
Single-cell 3C+ experimental approaches shed light on variation and regularities in chromatin patterns among individual cells 55. Single-cell 3C+ data can be processed with pairtools almost the same way as bulk Hi-C, with the addition of one filter. In single cells, the number of contacts produced by each genomic site is limited by the chromosome copy number. Thus, regions with irregularly high coverage indicate amplification artifacts or copy number variations 14 55 and must be excluded from downstream analyses 13 62. Pairtools filterbycov calculates genome-wide coverage per cell and excludes regions with coverage exceeding the specified threshold. This procedure helped to remove regions with anomalous coverage in single-cell Hi-C studies in Drosophila 14.
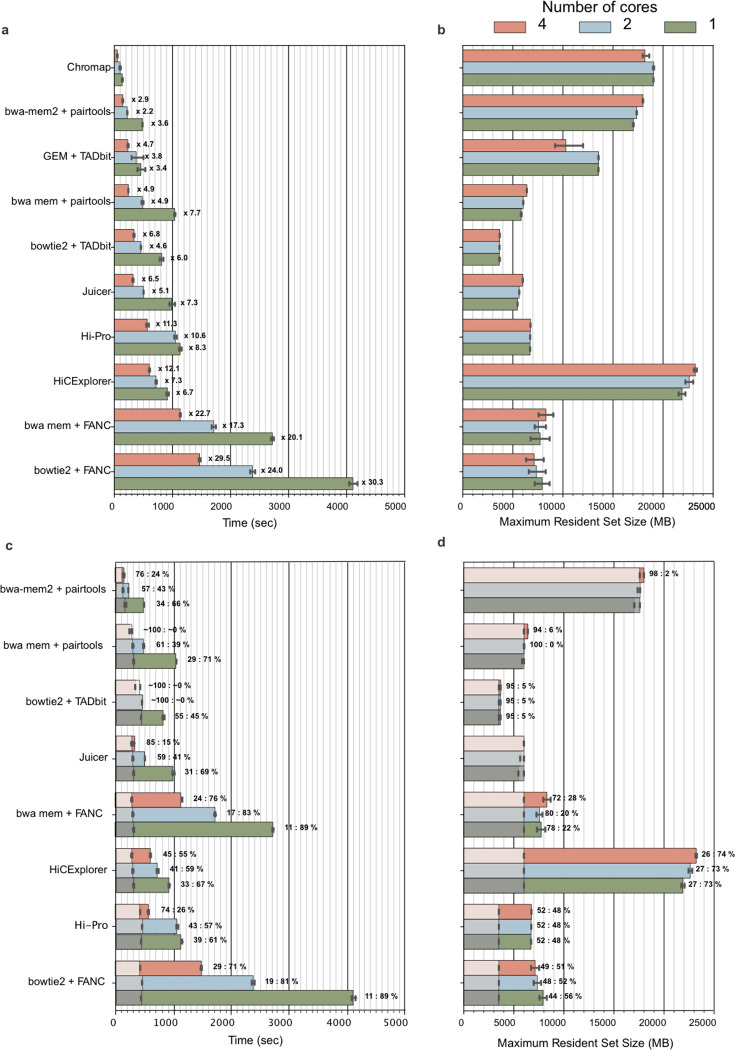
Performance and comparison with other tools
Contact extraction from raw sequencing data is the first and typically most time-consuming step of the 3C+ data processing. Pairtools is one of the fastest methods (lagging behind only Chromap 63) without consuming more memory (in combination with bwa mem), making it the best candidate for scalable 3C+ data processing (Figure 4). Notably, Pairtools is the only tool that combines high performance with the flexibility to enable adaptations to a broad range of 3C+protocols (Table 1).
Figure 4. Benchmark of different Hi-C mapping tools for one mln reads in 5 iterations (data from 39).
a. Runtime per tool and number of cores. The labels at each bar of the time plot indicate the slowdown relative to Chromap 63 with the same number of cores.
b. Maximum resident set size for each tool and number of cores.
c. Runtime per tool and number of cores compared to the runtime of the corresponding mapper (gray shaded areas). Labels at the bars reflect the percentage of time used by the mapper versus the time used by the pair parsing tool.
d. Maximum resident set size for each tool and number of cores compared with that of the corresponding mapper.
To make the comparison possible, the analysis for each tool starts with .fastq files, and the time includes both read alignment and pairs parsing. For pairtools, we tested the performance with regular bwa mem 29 and bwa mem2 30, which is ~2x faster but consumes more memory. Note that for HiC-Pro, we benchmark the original version and not the recently-rewritten nextflow 69 version that is part of nf-core 70. FANC, in contrast to other modular 3C+ pairs processing tools, requires an additional step to sort .bam files before parsing pairs that we include in the benchmark. For Juicer, we use the “early” mode. Chromap is not included in this comparison because it is an integrated mapper 63.
Table 1.
Qualitative comparison of the tools for pairs extraction from 3C+ sequencing data.
| Tool | Short description | Input | Output | Modular | Flexible | Aligner | Restriction sites | Quality control | Support for modified 3C+ protocols |
|---|---|---|---|---|---|---|---|---|---|
| Pairtools | python API/CLI tools | .sam alignments | pairs | Yes | Yes | bwa mem, bwa mem2, minimap2, bwa aln | Yes | Aggregated stats, scaling | Haplotype-resolved; chemical modification-based; multi-contact; single-cell |
| Chromap 63 | single executable | .fastq reads | contact maps | No | No | chromap | No | No | No |
| Juicer 64 | java/shell script pipeline | .fastq reads | pairs and contact maps | No | Yes | bwa mem | Yes | Aggregated stats | Haplotype-resolved |
| HiC-Pro 65 | python/R/shell script pipeline | .fastq reads | pairs and contact maps | No | Yes | bowtie2 | Yes | QC report | Haplotype-resolved |
| HiCExplorer 66 | python API/CLI tools | .sam alignments | contact maps | Yes * | Yes | bwa mem | Yes | QC report | No |
| FANC 67 | python API/CLI tools | .fastq reads or .sam alignments | pairs | Yes | Yes | bwa mem, bowtie2 | Yes | QC report | No |
| TADbit 68 | python API/CLI tools | .fastq reads or .sam alignments | parsed reads or contact maps | Yes | Yes | gem, bowtie2, hisat | Yes | Yes | No |
We consider methods modular if they have multiple tools that can be used separately or combined in a custom order. HiCExplorer is modular, but its tool for contact extraction is not (indicated with *). We consider methods flexible if they allow parameterization of data processing (e.g., restriction enzyme). We do not consider control only over technical parameters, like the number of cores, to be flexible. For restriction sites, we consider whether a method can either annotate or filter by restriction site.
Discussion
Pairtools provides a set of interoperable and high-performance utilities for processing 3C+ contact data 19 71 72, particularly for extraction of contacts from sequence alignments, manipulating pair files, deduplication, data filtering, generation of summary statistics and contact scalings, quality control, and treatment of data generated with 3C+ protocol modifications.
pairtools is easy to install via pip and conda. We provide extensive documentation of pairtools 73 , including example scripts of minimal pairtools-based pipelines and Jupyter tutorials with explanations of the working of pairtools, restriction, and phasing in pairtools GitHub repository 74.
The modular structure of pairtools and its usage of the .pairs format 28 already make it useful in many pipelines. pairtools is used in the 4DN pipeline (standard Hi-C) 3, the PORE-C pipeline (multi-way Hi-C) 75, HI-CAR nf-core pipeline (open-chromatin-associated contacts) 76 , and iMARGI pipelines (RNA-DNA contacts) 49 77. pairtools also serve as the foundation of distiller 78, a feature-rich fastq-to-cooler 18 pipeline, based on the nextflow workflow framework 79 and maintained by Open2C 72. Distiller takes advantage of piping pairtools command outputs and can parallelize 3C+ data processing within a single machine, on a cluster, or in the cloud.
In the future, a binary storage format for pairs could substantially speed up 3C+ contact extraction. Currently, zarr 80 is the best candidate as it allows variable length strings (not supported by hdf5 81) and allows appending columns and storing multiple tables in a single file (not supported by parquet 82).
To summarize, Pairtools provides an adaptable framework for future development to enable the expanding universe of 3C+ protocols.
Supplementary Material
Acknowledgements
The authors thank Leonid Mirny, Job Dekker and members of the Center for 3D Structure and Physics of the Genome for feedback on tool functionality. AAG, NA and SV acknowledge support from the Center for 3D Structure and Physics of the Genome, funded by the National Institutes of Health Common Fund 4D Nucleome Program UM1-HG011536. AAG was partially supported by grant 17-00-00180, IMBA and the Austrian Academy of Sciences (OeAW) during the early development of the project. GF is supported by R35 GM143116-01. AG is supported by IMBA and the Austrian Academy of Sciences (OeAW).
Footnotes
Code availability Statement
Open-source code is freely available at https://github.com/open2c/pairtools. Additional documentation is available at https://pairtools.readthedocs.io/, with interactive tutorials.
Pairtools is integrated into the high-performant nextflow-based pipeline distiller:
https://github.com/open2c/distiller-nf/.
Code for generating manuscript figures available at:
https://github.com/open2c/open2c_vignettes/tree/main/pairtools_manuscript/. Benchmark is available at https://github.com/open2c/pairtools/tree/master/doc/examples/benchmark/.
References
- 1.Akgol Oksuz B, Yang L, Abraham S, Venev SV, Krietenstein N, Parsi KM et al. Systematic evaluation of chromosome conformation capture assays. Nat Methods 2021; 18: 1046–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S et al. The 4D nucleome project. Nature 2017; 549: 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo Y, Hitz BC, Gabdank I, Hilton JA, Kagda MS, Lam B et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res 2020; 48: D882–D889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan H, Onichtchouk D, Winata C. DANIO-CODE: Toward an Encyclopedia of DNA Elements in Zebrafish. Zebrafish 2016; 13: 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh T-HS, Fudenberg G, Goloborodko A, Rando OJ. Micro-C XL: assaying chromosome conformation from the nucleosome to the entire genome. Nat Methods 2016; 13: 1009–1011. [DOI] [PubMed] [Google Scholar]
- 7.Lafontaine DL, Yang L, Dekker J, Gibcus JH. Hi-C 3.0: Improved protocol for genome-wide chromosome conformation capture. Curr Protoc 2021; 1: e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erceg J, AlHaj Abed J, Goloborodko A, Lajoie BR, Fudenberg G, Abdennur N et al. The genome-wide multi-layered architecture of chromosome pairing in early Drosophila embryos. Nat Commun 2019; 10: 4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitter M, Takacs Z, Köcher T, Micura R, Langer CCH, Gerlich DW. Sister chromatid–sensitive Hi-C to map the conformation of replicated genomes. Nat Protoc 2022; 17: 1486–1517. [DOI] [PubMed] [Google Scholar]
- 10.Oomen ME, Hedger AK, Watts JK, Dekker J. Detecting chromatin interactions between and along sister chromatids with SisterC. Nat Methods 2020; 17: 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013; 502: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano T, Lubling Y, Várnai C, Dudley C, Leung W, Baran Y et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 2017; 547: 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 2017; 544: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulianov SV, Zakharova VV, Galitsyna AA, Kos PI, Polovnikov KE, Flyamer IM et al. Order and stochasticity in the folding of individual Drosophila genomes. Nat Commun 2021; 12: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan L, Ma W, Wu H, Zheng Y, Xing D, Chen R et al. Changes in genome architecture and transcriptional dynamics progress independently of sensory experience during post-natal brain development. Cell 2021; 184: 741–758.e17. [DOI] [PubMed] [Google Scholar]
- 16.Deshpande AS, Ulahannan N, Pendleton M, Dai X, Ly L, Behr JM et al. Identifying synergistic high-order 3D chromatin conformations from genome-scale nanopore concatemer sequencing. Nat Biotechnol 2022. doi: 10.1038/s41587-022-01289-z. [DOI] [PubMed] [Google Scholar]
- 17.Tavares-Cadete F, Norouzi D, Dekker B, Liu Y, Dekker J. Multi-contact 3C reveals that the human genome during interphase is largely not entangled. Nat Struct Mol Biol 2020; 27: 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdennur N, Mirny LA. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 2020; 36: 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Open2C, Abdennur N, Abraham S, Fudenberg G, Flyamer IM, Galitsyna AA et al. Cooltools: enabling high-resolution Hi-C analysis in Python. bioRxiv. 2022; : 2022.10.31.514564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D et al. Array programming with NumPy. Nature 2020; 585: 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinney W. Data Structures for Statistical Computing in Python. In: Proceedings of the 9th Python in Science Conference. SciPy, 2010. doi:10.25080/majora-92bf1922-00a. [Google Scholar]
- 22.McIlroy MD, Pinson EN, Tague BA. UNIX time-sharing system: Foreword. The Bell System Technical Journal 1978; 57: 1899–1904. [Google Scholar]
- 23.Mckinney W. Pandas: A foundational Python library for data analysis and statistics.https://www.dlr.de/sc/portaldata/15/resources/dokumente/pyhpc2011/submissions/pyhpc2011_submission_9.pdf (accessed 24 Oct2022). [Google Scholar]
- 24.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 2020; 17: 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011; 12: 2825–2830. [Google Scholar]
- 26.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO et al. Twelve years of SAMtools and BCFtools. Gigascience 2021; 10. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S, Bakker C, Vitzthum C, Alver BH, Park PJ. Pairs and Pairix: a file format and a tool for efficient storage and retrieval for Hi-C read pairs. Bioinformatics 2022. doi: 10.1093/bioinformatics/btab870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bio.GN]. 2013.http://arxiv.org/abs/1303.3997. [Google Scholar]
- 30.Vasimuddin M, Misra S, Li H, Aluru S. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS). 2019. doi: 10.1109/ipdps.2019.00041. [DOI] [Google Scholar]
- 31.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018; 34: 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krietenstein N, Abraham S, Venev SV, Abdennur N, Gibcus J, Hsieh T-HS et al. Ultrastructural details of mammalian chromosome architecture. doi: 10.1101/639922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang L-H, Ghosh S, Papale A, Miranda M, Piras V, Degrouard J et al. A complex CTCF binding code defines TAD boundary structure and function. bioRxiv. 2021; : 2021.04.15.440007. [Google Scholar]
- 34.Jiang T, Raviram R, Snetkova V, Rocha PP, Proudhon C, Badri S et al. Identification of multi-loci hubs from 4C-seq demonstrates the functional importance of simultaneous interactions. Nucleic Acids Res 2016; 44: 8714–8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oudelaar AM, Harrold CL, Hanssen LLP, Telenius JM, Higgs DR, Hughes JR. A revised model for promoter competition based on multi-way chromatin interactions at the α-globin locus. Nat Commun 2019; 10: 5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dotson GA, Chen C, Lindsly S, Cicalo A, Dilworth S, Ryan C et al. Deciphering multi-way interactions in the human genome. Nat Commun 2022; 13: 5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olivares-Chauvet P, Mukamel Z, Lifshitz A, Schwartzman O, Elkayam NO, Lubling Y et al. Capturing pairwise and multi-way chromosomal conformations using chromosomal walks. Nature 2016; 540: 296–300. [DOI] [PubMed] [Google Scholar]
- 38.Hsieh T-HS, Cattoglio C, Slobodyanyuk E, Hansen AS, Darzacq X, Tjian R. Enhancer-promoter interactions and transcription are maintained upon acute loss of CTCF, cohesin, WAPL, and YY1. bioRxiv. 2021; : 2021.07.14.452365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao SSP, Huang S-C, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon K-R et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017; 171: 305–320.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du Q, Smith GC, Luu PL, Ferguson JM, Armstrong NJ, Caldon CE et al. DNA methylation is required to maintain both DNA replication timing precision and 3D genome organization integrity. Cell Rep 2021; 36: 109722. [DOI] [PubMed] [Google Scholar]
- 41.Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics 2011; 27: 718–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bentley JL. Multidimensional binary search trees used for associative searching. Commun ACM 1975; 18: 509–517. [Google Scholar]
- 43.Picard. http://broadinstitute.github.io/picard/ (accessed 30 Jan2023).
- 44.Thread: [Samtools-help] Pickard estimate for the size of a library - wrong or non-transparent? https://sourceforge.net/p/samtools/mailman/samtools-help/thread/DUB405-EAS154589A1ACEF2BE4C573D4592180@phx.gbl/ (accessed 30 Jan2023).
- 45.Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods 2012; 9: 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014; 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galitsyna AA, Khrameeva EE, Razin SV, Gelfand MS. Mirror reads. Hi-C data Genomics and. [Google Scholar]
- 48.Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA et al. Organization of the mitotic chromosome. Science 2013; 342: 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu W, Yan Z, Nguyen TC, Bouman Chen Z, Chien S, Zhong S. Mapping RNA–chromatin interactions by sequencing with iMARGI. Nat Protoc 2019; 14: 3243–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Library QC — micro-C 0.1 documentation. https://micro-c.readthedocs.io/en/latest/library_qc.html (accessed 24 Oct2022).
- 51.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016; 32: 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goel VY, Hansen AS. The macro and micro of chromosome conformation capture. Wiley Interdiscip Rev Dev Biol 2021; 10: e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee D-S, Luo C, Zhou J, Chandran S, Rivkin A, Bartlett A et al. Simultaneous profiling of 3D genome structure and DNA methylation in single human cells. Nat Methods 2019; 16: 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li G, Liu Y, Zhang Y, Kubo N, Yu M, Fang R et al. Joint profiling of DNA methylation and chromatin architecture in single cells. Nat Methods 2019; 16: 991–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galitsyna AA, Gelfand MS. Single-cell Hi-C data analysis: safety in numbers. Brief Bioinform 2021; 22. doi: 10.1093/bib/bbab316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lajoie BR, Dekker J, Kaplan N. The Hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods 2015; 72: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramani V, Cusanovich DA, Hause RJ, Ma W, Qiu R, Deng X et al. Mapping 3D genome architecture through in situ DNase Hi-C. Nat Protoc 2016; 11: 2104–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.AlHaj Abed J, Erceg J, Goloborodko A, Nguyen SC, McCole RB, Saylor W et al. Highly structured homolog pairing reflects functional organization of the Drosophila genome. Nat Commun 2019; 10: 4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng H, Jarvis ED, Fedrigo O, Koepfli K-P, Urban L, Gemmell NJ et al. Haplotype-resolved assembly of diploid genomes without parental data. Nat Biotechnol 2022; 40: 1332–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collombet S, Ranisavljevic N, Nagano T, Varnai C, Shisode T, Leung W et al. Parental-to-embryo switch of chromosome organization in early embryogenesis. Nature 2020; 580: 142–146. [DOI] [PubMed] [Google Scholar]
- 61.Tan L, Xing D, Chang C-H, Li H, Xie XS. Three-dimensional genome structures of single diploid human cells. Science 2018; 361: 924–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gassler J, Brandão HB, Imakaev M, Flyamer IM, Ladstätter S, Bickmore WA et al. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J 2017; 36: 3600–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang H, Song L, Wang X, Cheng H, Wang C, Meyer CA et al. Fast alignment and preprocessing of chromatin profiles with Chromap. Nat Commun 2021; 12: 6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst 2016; 3: 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Servant N, Varoquaux N, Lajoie BR, Viara E, Chen C-J, Vert J-P et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 2015; 16: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolff J, Bhardwaj V, Nothjunge S, Richard G, Renschler G, Gilsbach R et al. Galaxy HiCExplorer: a web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res 2018; 46: W11–W16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kruse K, Hug CB, Vaquerizas JM. FAN-C: a feature-rich framework for the analysis and visualisation of chromosome conformation capture data. Genome Biol 2020; 21: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Serra F, Baù D, Goodstadt M, Castillo D, Filion GJ, Marti-Renom MA. Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLoS Comput Biol 2017; 13: e1005665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 2020; 38: 276–278. [DOI] [PubMed] [Google Scholar]
- 70.Servant N, Peltzer A. nf-core/hic: Initial release of nf-core/hic. 2019. doi: 10.5281/zenodo.2669513. [DOI] [Google Scholar]
- 71.Open2C, Abdennur N, Fudenberg G, Flyamer I, Galitsyna AA, Goloborodko A et al. Bioframe: Operations on Genomic Intervals in Pandas Dataframes. bioRxiv. 2022; : 2022.02.16.480748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Welcome to the Open Chromosome Collective! Open2C. https://open2c.github.io/ (accessed 1 Nov2022).
- 73.Overview — pairtools 1.0.2 documentation. http://pairtools.readthedocs.io/en/latest/ (accessed 24 Oct2022).
- 74.pairtools: CLI tools to process mapped Hi-C data. Github https://github.com/open2c/pairtools (accessed 24 Oct2022).
- 75.Pore-C-Snakemake. Githubhttps://github.com/nanoporetech/Pore-C-Snakemake (accessed 24 Oct2022).
- 76.Wei X, Xiang Y, Peters DT, Marius C, Sun T, Shan R et al. HiCAR is a robust and sensitive method to analyze open-chromatin-associated genome organization. Mol Cell 2022; 82: 1225–1238.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Overview — iMARGI pipeline 1.1 documentation. http://sysbiocomp.ucsd.edu/public/frankyan/imargi_pipeline/ (accessed 24 Oct2022). [Google Scholar]
- 78.Goloborodko A, Venev S, Abdennur N, Tommaso PD. mirnylab/distiller-nf: v0. 3.3. 2019. [Google Scholar]
- 79.Di Tommaso P, Chatzou M, Floden EW, Barja PP, Palumbo E, Notredame C. Nextflow enables reproducible computational workflows. Nat Biotechnol 2017; 35: 316–319. [DOI] [PubMed] [Google Scholar]
- 80.Zarr — zarr 2.13.3 documentation. https://zarr.readthedocs.io/en/stable/ (accessed 24 Oct2022). [Google Scholar]
- 81.The HDF5® Library & File Format. The HDF Group. 2017.https://www.hdfgroup.org/HDF5/ (accessed 24 Oct2022).
- 82.parquet-format: Apache Parquet. Githubhttps://github.com/apache/parquet-format (accessed 24 Oct2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.