Figure 2. CpG methylation gradients traverse chr4q and 10q D4Z4 arrays.
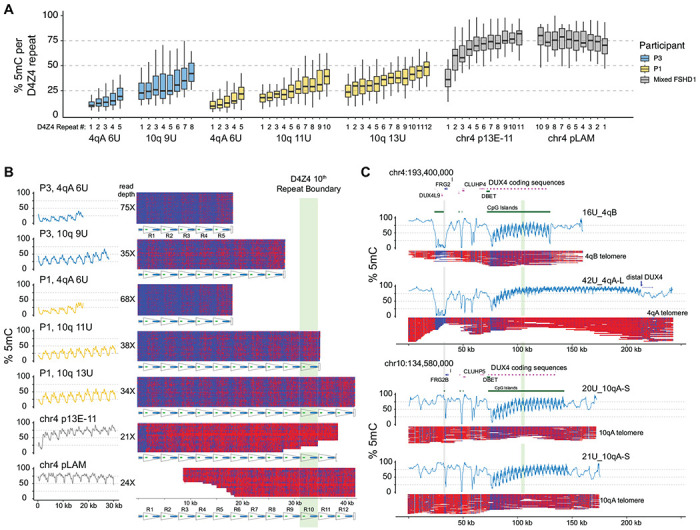
(A) Methylation levels of p13E-11 to pLAM targeted reads summarized as % 5mC sites per D4Z4 repeat (3.3 kb KpnI-to-KpnI region with ~320 CpG sites per repeat). Repeat numbering begins at the centromeric end (p13E-11) and is reversed for the chr4 pLAM (telomeric) reads. (B) Single-read plots of targeted reads from (A) using modbamtools with unmethylated CpGs represented in blue and 5-methyl CpGs in red. The schematic representation of D4Z4 repeats shows DUX4 exons 1 and 2 and the CTCF insulator region, and the green stripe indicates the 10th centromeric D4Z4 repeat. (C) Alignments and methylation plots of untargeted reads (R10.4.1_e8.2 nanopores) from a control subject (C4) mapped to the 4qB 16U haplotype, the 4qA 43U haplotype, the 10q 20U haplotype, and the 10q 21U haplotype. The grey stripe delineates the start of the syntenic telomeric region shared between 4q and 10q, and the green stripe marks the 10th repeat.
