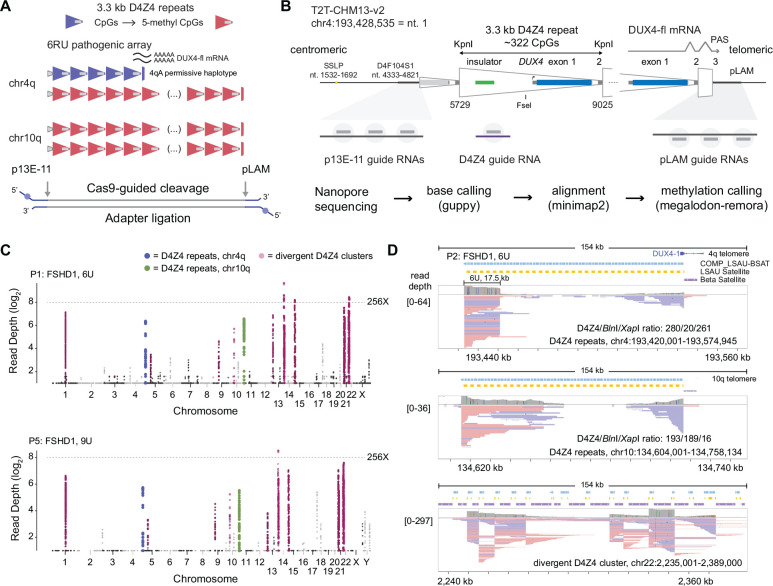
Figure 1. Cas9-targeted nanopore sequencing of D4Z4 repeats.
(A) Schematic of D4Z4 repeat arrays found in sub-telomeric regions of human chr4q and 10q. A 6U D4Z4 array is shown on the permissive 4qA haplotype, and blue/red shading of repeats indicates CpG methylation. (B) Locations of Cas9-guide RNA cleavage sites at the centromeric (p13E-11) end, telomeric (pLAM) end, and within each repeat (D4Z4). Steps used in the sequencing pipeline for identifying targeted reads are shown. (C) Read depth Manhattan plots from two FSHD1 participants (P1, P5) mapped to the T2T CHM13 v2.0 reference genome (log2 scale, minimum depth = 2 reads, summarized in 2 kb, non-overlapping windows). Colors indicate reads mapped to the chr4q (blue) and 10q (green) D4Z4 arrays, as well as divergent DUX4 clusters (purple). (D) IGV browser alignments of targeted reads from FSHD1 participant P5 with a 4qA 6U contraction. The D4Z4/BlnI/XapI ratio shows the number of 3.3 kb D4Z4 repeats contained with the aligned reads and the number of 10q-specific (BlnI) and 4q-specific (XapI) restriction sites found within those repeats. The lower panel shows alignments to a divergent D4Z4 repeat cluster on chr22.