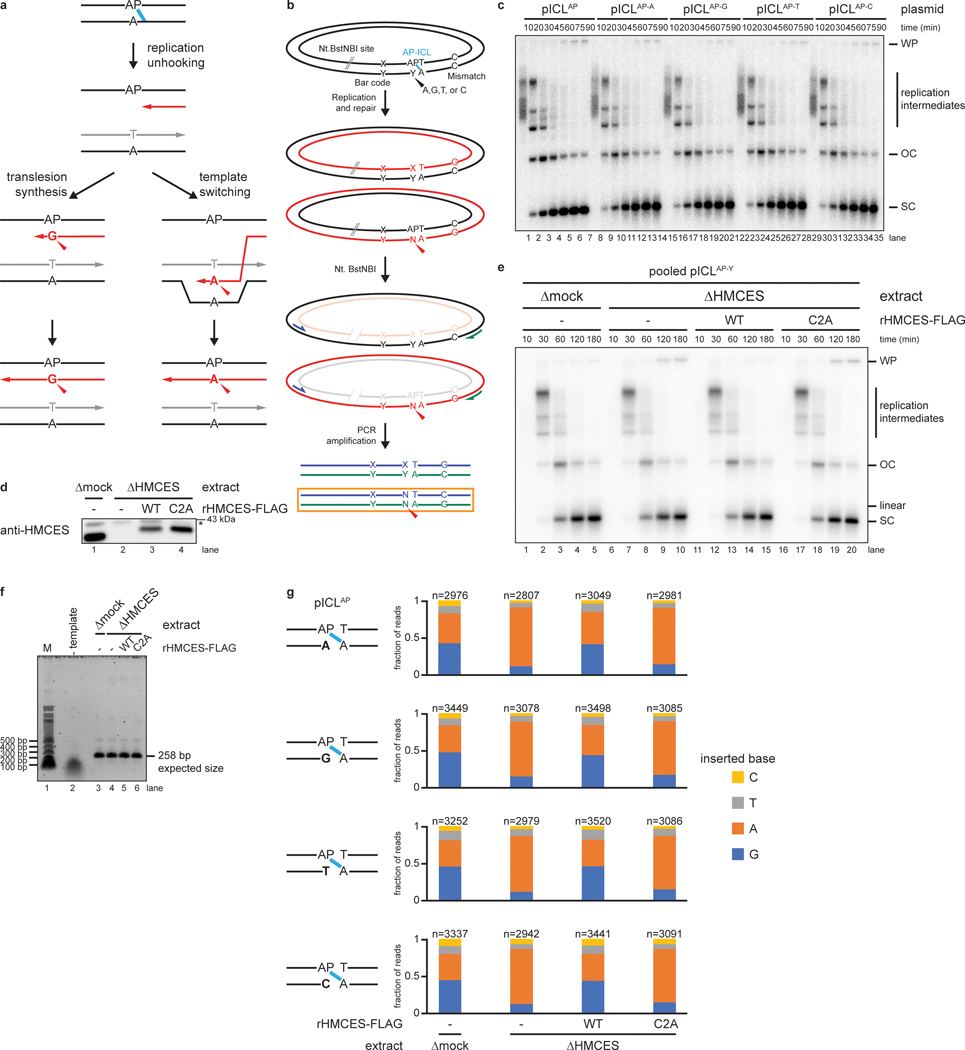
Extended Data Fig. 6. Nucleotide identity opposite the AP site does not effect NEIL3-dependent ICL unhooking or TLS.
a, Model of two alternative mechanisms of AP site bypass. Translesion synthesis (left branch) uses a specialized TLS polymerase for untemplated insertion of a nucleotide opposite the non-coding AP site. In this pathway, the nucleotide opposite the AP site in the parental plasmid does not influence insertion by the TLS polymerase. Template switching (right branch) uses the newly synthesized DNA of the other sister chromatid as a template for error-free bypass of the AP site. In this case, the inserted nucleotide is expected to co-vary with the nucleotide opposite the AP site in the parental plasmid.
b, Plasmid design for next generation sequencing. We prepared four different plasmids, each containing an AP-ICL and a different nucleotide positioned opposite the AP site (Y), as well as a unique barcode (X-Y). An Nt.BstNBI restriction site allows specific cleavage of the AP site-containing strand to prevent its amplification during PCR. A dC-dC mismatch allows reads produced from amplification of the nascent strand that has bypassed the AP site (PCR product in orange box) to be distinguished from reads derived from the corresponding parental strand (upper PCR product).
c, The four AP-ICL plasmids, each containing a different nucleotide opposite the AP-site (described in b), were replicated in undepleted egg extract supplemented with [α−32P]dATP. Replication intermediates were analyzed as in Fig. 2a. The base opposite the AP site had no apparent effect on replication or repair efficiency.
d, HMCES immunodepletion. The extracts used to generate the sequencing libraries described in Fig. 5 were blotted for HMCES. Asterisk, non-specific band.
e, The four AP-ICL plasmids described in b were pooled and replicated with [α−32P]dATP in the same extracts (shown in panel d) and used to generate sequencing libraries described in Fig. 5. Replication intermediates were analyzed as in Fig. 2a.
f, Analysis of PCR amplicons used for sequencing. In parallel to the reactions shown in e, pooled pICLAP plasmids were replicated in the indicated extracts (shown in d, but lacking [α−32P]dATP) supplemented with rHMCES, as indicated. DNA was extracted and digested with Nt.BstNBI (to cleave AP site-containing strands). The region of the replicated plasmids surrounding the ICL was then amplified by PCR. PCR amplicons were resolved by native agarose gel electrophoresis and visualized by Sybr Gold staining.
g, Analysis of sequencing reads derived from individual pICLAP plasmids. The barcode was used to distinguish sequencing reads derived from the four different AP-ICL containing plasmids described in b. For each extract condition, we obtained >30,000 mapped reads, the vast majority (87.4%−87.8%) of which either perfectly matched the reference sequence or had a single point mutation corresponding to the position opposite the AP site. Of these reads, >11,000 in each condition derived from the nascent DNA stand produced upon bypass of the AP site. The fraction of reads corresponding to insertion of a given nucleotide opposite the AP site are plotted for each plasmid and extract condition. n, number of pooled nascent strand reads obtained for each condition. The result shows that the nucleotide opposite the AP site in the parental plasmid template does not influence the distribution of nascent DNA nucleotides inserted opposite the AP site after unhooking. Next generation sequencing read counts can be found in Supplementary Table 2.