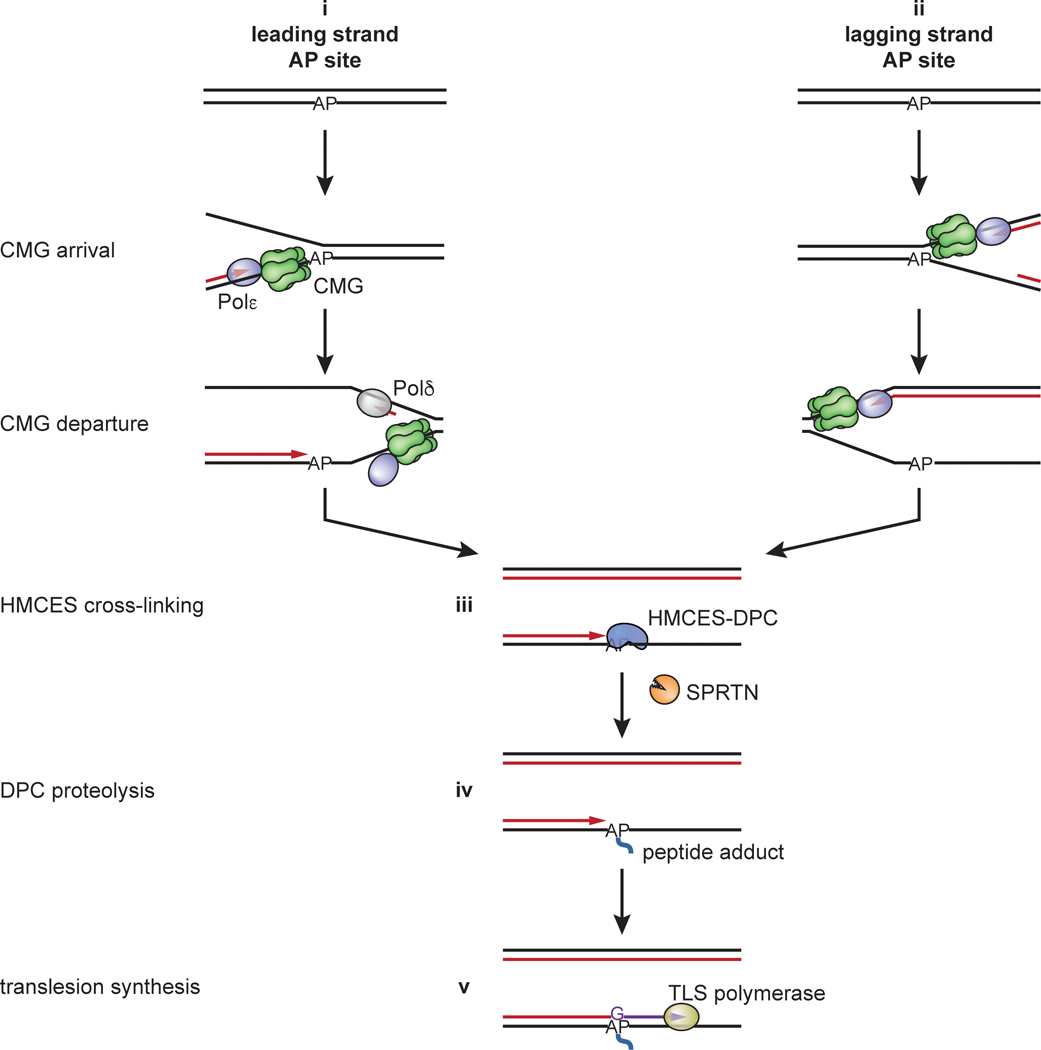
Extended Data Fig. 9. A proposed general model for AP site protection by HMCES during DNA replication.
(i) AP sites encountered in the leading strand template due to spontaneous depurination/depyrimidination or incomplete BER are bypassed by CMG. This leads to uncoupling of DNA unwinding by CMG from leading strand DNA synthesis by Pol ε and HMCES-DPC formation at a ssDNA/dsDNA junction. (ii) AP sites in the lagging strand template are bypassed without CMG uncoupling, and HMCES cross-links to the AP site fully embedded in ssDNA. In both scenarios, Pol δ then extends the lagging strand up to the HMCES-DPC, where upon synthesis stalls (iii). In both cases, the HMCES-DPC abuts a ssDNA/dsDNA junction, leading to proteolysis by SPRTN (iv). The HMCES-DPC and the peptide adduct generated after proteolysis stabilize the AP site until the lesion is bypassed by TLS (v) or an alternative, error-free mechanism (not depicted).