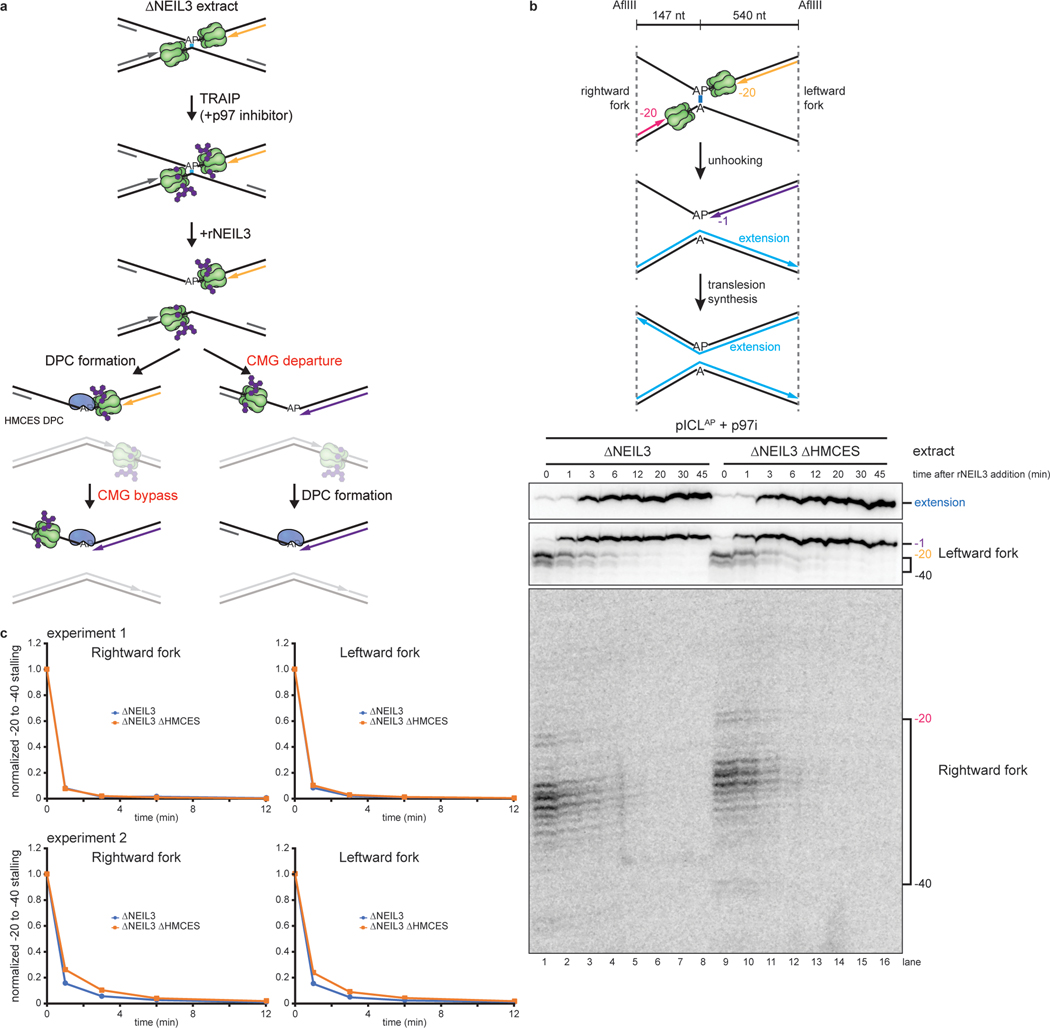
Fig. 3: HMCES-DPC formation does not impede CMG translocation.
a, Models for timing of HMCES-DPC formation. Left branch, HMCES-DPC formation precedes CMG bypass of the AP site, which is predicted to delay rightward leading strand approach to the AP site. Right branch, CMG bypass of AP site precedes HMCES-DPC formation, resulting in no delay in rightward leading strand approach.
b, Top, Schematic of nascent strands generated during ICL repair. AflIII cuts 147 nucleotides to the left and 540 nucleotides to the right of the ICL, respectively, generating characteristic −20 to −40 stall, −1 stall, and strand extension products. Bottom, pICLAP was replicated with [α−32P]dATP and p97i in the indicated extracts (shown in Extended Data Fig. 4a). After 60 minutes, rNEIL3 was added and nascent DNA strands were isolated at the indicated times, digested with AflIII, and resolved by denaturing polyacrylamide gel electrophoresis. Top, middle, and bottom panels show sections of the same gel to visualize extension, leftward leading strands, and rightward leading strands, respectively. Contrast was adjusted to visualize rightward fork stall products.
c, The persistence of −20 to −40 stall products in b was quantified by dividing the summed intensities of the −20 to −40 stall product bands in each lane by the intensity of the rightward fork −1 stall product band. Quantifications were normalized to the accumulation of −20 to −40 stall products at the 0 min time time point relative to NEIL3 addition. Quantifications from two independent experiments are shown.