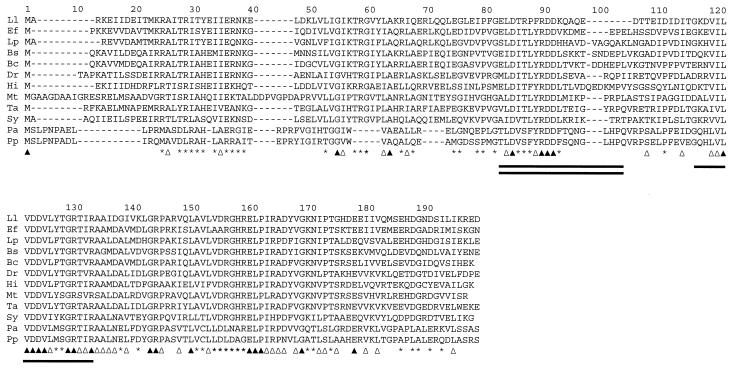
FIG. 6.
Clustal W alignment of amino acid sequences deduced from pyrR homologues of L. lactis (Ll), E. faecalis (Ef), L. plantarum (Lp), B. subtilis (Bs), B. caldolyticus (Bc), Deinococcus radiodurans (Dr), Haemophilus influenzae (Hi), Mycobacterium tuberculosis (Mt), Thermus aquaticus (Ta), Synechocystis sp. (Sy), Pseudomonas aeruginosa (Pa), and Pseudomonas putida (Pp). The sequences were retrieved from protein databases after a Blast search using the search engine at the National Center for Biotechnology Information. Solid triangles, amino acid residues conserved in all sequences; open triangles, positions where only conservative substitutions have occurred; asterisks, positions where the amino acid residues are conserved in more than 66% of sequences; single line, putative binding site for PRPP; double line, flexible-loop domain.