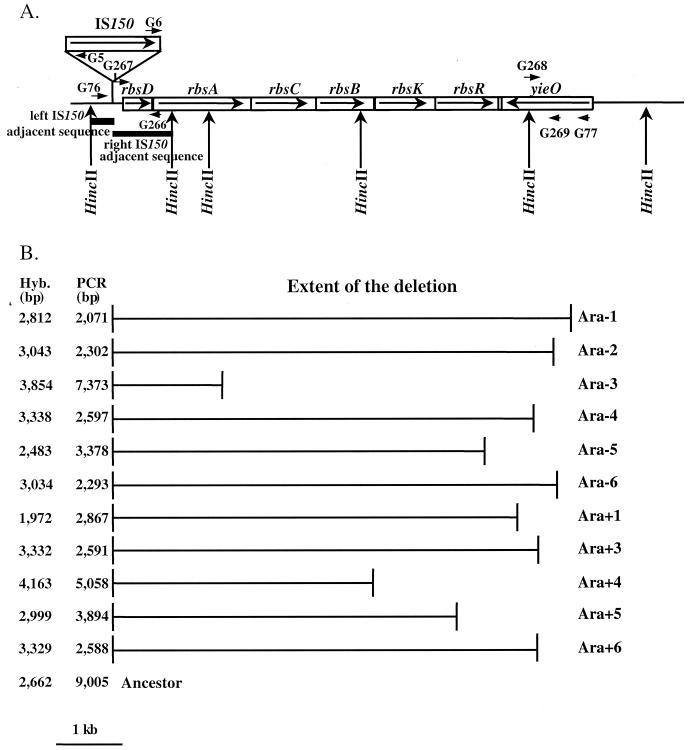
FIG. 3.
Losses of d-ribose catabolism in evolving populations caused by deletions in the rbs operon. (A) Map of the rbs operon in E. coli B ancestor, based on the genome sequence for E. coli K-12 (4). Arrows show the directions of gene transcription. An IS150 element is located upstream of the rbs operon. Thick lines indicate the left and right adjacent sequences used as probes. The primers used in PCR experiments (G5, G6, G76, and G77) and for constructing strain GBE127 (G266, G267, G268, and G269) are also shown, as are relevant HincII restriction sites. (B) The deletions in clones isolated from 11 of the 12 populations are shown as horizontal lines. Population Ara+2 is not depicted because an Rbs+ minority clone was sampled (see text). The right adjacent sequence of IS150 did not hybridize with any of the Rbs− clones, whereas the left adjacent sequence hybridized in every case with a HincII fragment, whose size is shown in the column labeled Hyb. The sizes of the PCR products obtained from these clones using primers G76 and G77 are given in the column labeled PCR. The sizes, in the ancestral strain, of the hybridizing HincII fragment and PCR product are indicated at the bottom.