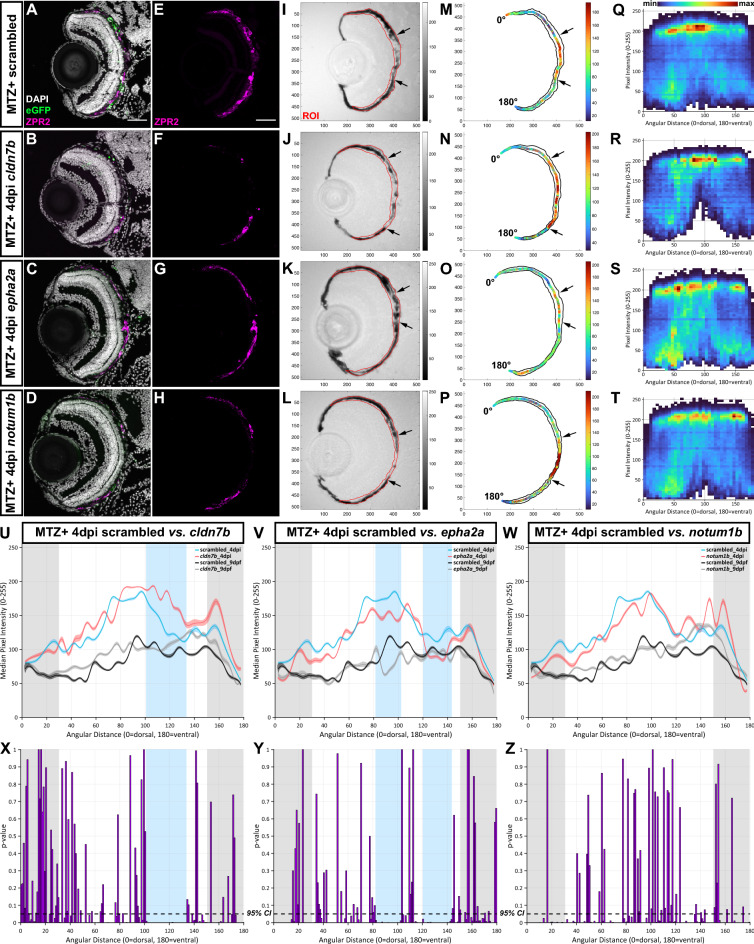
Figure 2.
Representative screening results. (A–L) Representative immunofluorescence images of transverse cryosections from ablated (MTZ+) scrambled, cldn7b, epha2a, and notum1b RNP-injected F0 larvae at 4dpi. Single-channel ZPR2 (E–H) and brightfield (I–L) images. Grayscale bars indicate an 8-bit scale (0 = black, 255 = white). Black arrows delineate edges of pigment recovery. Regions of interest (ROIs) are outlined in red. Nuclei (white), eGFP (green), ZPR2 (magenta). (M–P) Median pixel intensity distributions in the ROI. Color bars indicate an 8-bit scale (0 = dark blue, 255 = dark red). (Q–T) Heatmaps showing dorsal-to-ventral (angular distance from 0 to 180°) distribution of raw pixel intensity values within each ROI from ablated scrambled, cldn7b, epha2a, and notum1b RNP-injected F0 larvae at 4dpi. Color bar above (Q) indicates bin counts. (U–W) Plots showing median ROI pixel intensity values from 4dpi ablated (blue and red lines) and 9dpf unablated (black and gray lines) scrambled, cldn7b, epha2a, and notum1b RNP-injected F0 larvae. (Q–W) Bin size = 5 angular degrees. (X–Z) Statistical comparisons of dorsal-to-ventral median pixel intensity between: (X) scrambled and cldn7b, (Y) scrambled and epha2a, and (Z) scrambled and notum1b. Blue shading indicates regions with significant differences spanning > 20 angular degrees. Gray shading indicates the dorsal (0 to 30 angular degrees) and ventral (150 to 180 angular degrees) peripheral RPE areas omitted from analyses. Dashed black lines indicate a 95% confidence interval (CI), Bin size = 1 angular degree. Scale bars = 50 μm. Exact regions with significant differences compared to scrambled controls can be found in Table S2Abbreviations as follows: MTZ Metronidazole; dpi Days post-injury; min Minimum; max Maximum.