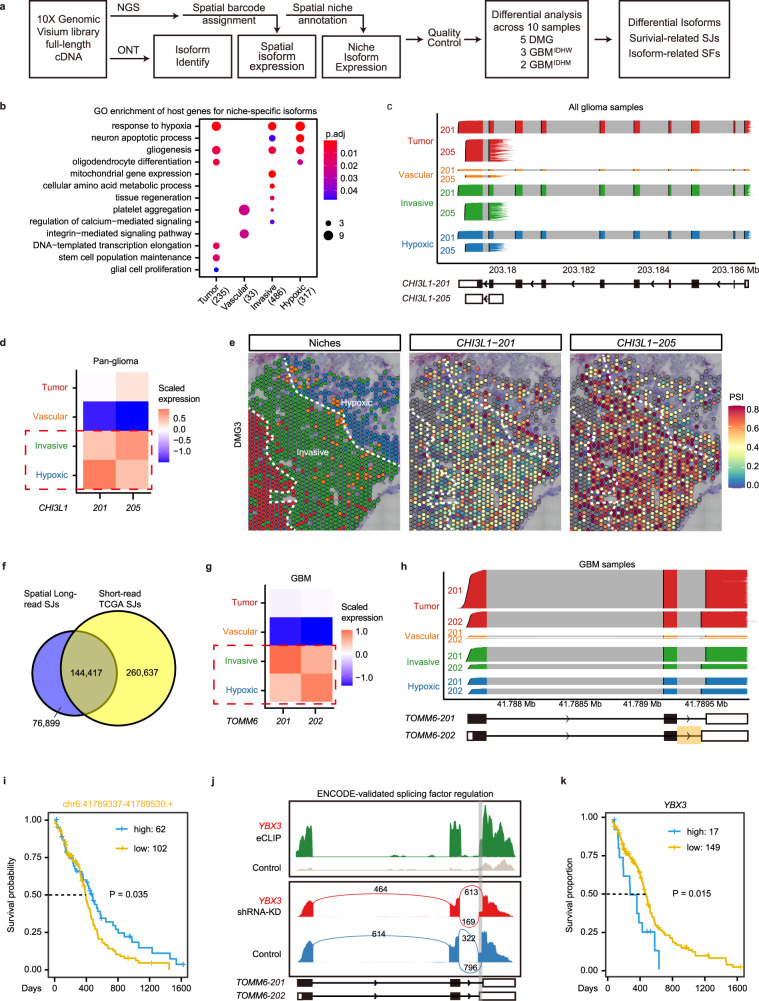
Fig. 3. RNA isoform diversity across glioma niches revealed by long-read spatial transcriptomics.
a Workflow to identify niche-specific isoforms and splicing junctions (SJs), predict regulatory splicing factors (SFs), and analyze their prognostic value for patient survival. b Gene Ontology (GO) enrichment of the host genes of niche-specific isoform across samples (n = 10). P values were determined by two tailed hypergeometric test. Significantly enriched (Benjamini–Hochberg-adjusted P < 0.05) biological processes are shown. c–e Differential isoform enrichment of CHI3L1 in different niches. c Individual transcripts of CHI3L1 isoforms 201 and 205 in high-quality malignant spots from all glioma samples (n = 19,071 spots) are represented by transcript tracks, colored by their niche identities. Transcript structures based on Ensembl annotation are shown at the bottom, with colored regions representing exons and gray regions representing introns. d Tile plots showing scaled mean expression of CHI3L1-201 and CHI3L1-205 across niches to compare their niche-specific enrichment patterns. e As an example, spatial plots of PSI values (colors) in DMG3 quantifying the relative expression of CHI3L1-201 and CHI3L1-205 are shown. f Venn diagram showing the overlap of splice junctions detected in our long reads sequencing dataset and the TCGA-GBM dataset37. Differential isoform enrichment of TOMM6-201 and TOMM6-202 in different niches of GBM samples (n = 6727 spots), shown by tile plots (g) and individual transcripts in each niche (h). i Kaplan–Meier survival curves comparing the overall survival in the TCGA-GBM cohort36,37, stratified by high (n = 62) vs. low expression (n = 102) of TOMM6-202 specific splice junction. Curve comparison P value was determined by two tailed Log-rank (Mantel–Cox) test. j Predicted splicing regulation of TOMM6 by splicing factor YBX3. Peaks indicate eCLIP and shRNA-KD RNA-seq read density in HepG2 cell line from ENCODE38. Biological replicates have similar results. Gray box shows the alternatively spliced region of TOMM6-202. k Kaplan–Meier survival curves comparing the overall survival between YBX3 high (n = 17) versus low groups (n = 149) in the TCGA-GBM cohort36. Curve comparison P value was determined by two tailed Log-rank (Mantel–Cox) test.