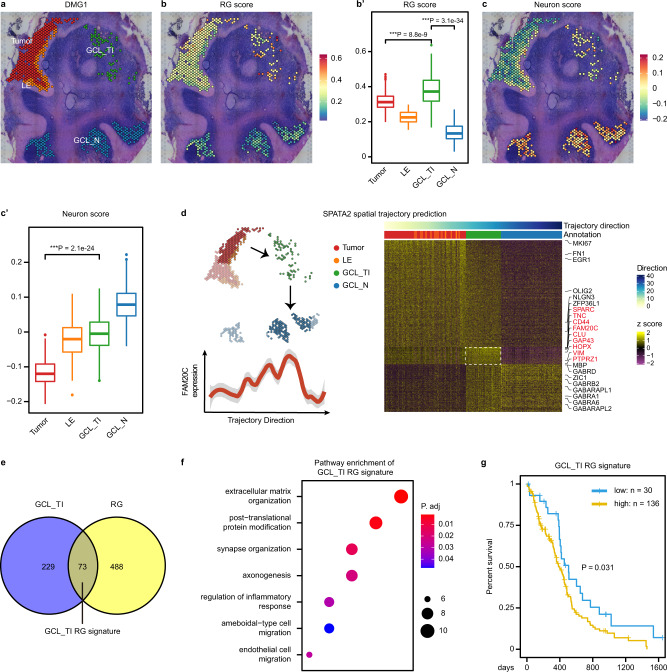
Fig. 5. Identification of regulatory programs of RG-like cells in a neuron-rich invasive niche.
a Selected spots from the tumor core (Tumor), leading edge (LE), and tumor-infiltrated GCL (GCL_TI), and normal spots without CNV from GCL (GCL_N) of DMG1 are shown, distinguished by color. b Spatial plot of the RG score in selected spots. b′ The RG score in each region (n = 256, 74, 71, and 200 spots, respectively) is quantified and compared. Boxes indicate quartiles, horizontal bar indicates median, and whiskers indicate range, up to 1.5-fold inter-quartile range. P values, Kruskal–Wallis test for between group differences with Holm’s correction for multiple comparisons. c Spatial plot of the Neuron score in selected spots. c′ The Neuron score in each region (n = 256, 74, 71, and 200 spots, respectively) is quantified and compared. Boxes indicate quartiles, horizontal bar indicates median, and whiskers indicate range, up to 1.5-fold inter-quartile range. P values, two tailed Wilcox test for between group differences for comparisons. d The spatial trajectory from Tumor to GCL_N inferred by SPATA243. Highlighted spots were selected for analysis (upper left). Heatmap of genes (row) dynamically expressed in selected spots (column, marked by different colors representing regions) along the spatial trajectory is shown on the right, clustered into three modules. Genes upregulated in GCL_TI exhibiting an ‘one-peak’ pattern is highlighted in red. The expression of FAM20C along the spatial trajectory is shown as an example (bottom left). The shaded area represents the 95% confidence interval. e Venn chart showing the overlap between genes upregulated in GCL_TI (d) and the RG signature genes. The 73 overlapping genes are termed GCL_TI RG signature. f Gene Ontology enrichment analysis of the GCL_TI RG signature genes. P value was determined by two tailed hypergeometric test. Top enriched (Benjamini–Hochberg-adjusted P < 0.05) biological processes are shown. g Kaplan–Meier curves comparing the overall survival between GCL_TI RG signature high (n = 136) versus low (n = 30) groups in the TCGA-GBM cohort. Curve comparison p value was determined by two tailed Log-rank (Mantel–Cox) test.