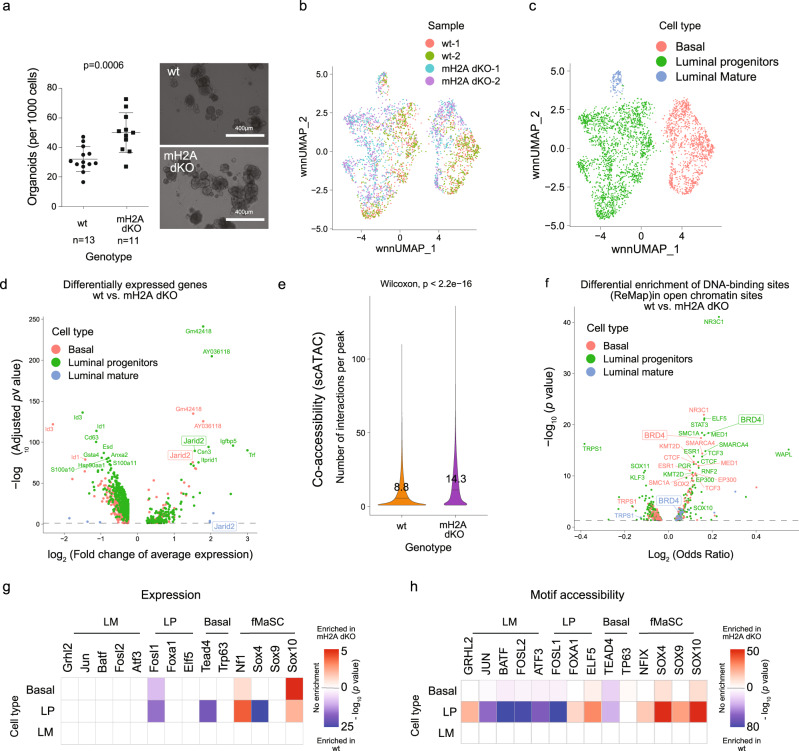
Fig. 7. Loss of mH2A histone variants in MaSC reveals a proto-oncogenic signature associated with increased Sox10 activity.
a Number of organoids per 100 cells plated from wt and mH2A dKO mouse mammary epithelial cells after 7 days in culture. Data are mean with SD (p = 0.0006, unpaired t-test). b Weighted nearest neighbor (WNN) UMAP plot—combining single cell gene expression and single cell ATAC-seq signals, of mammary stem cells from wild type (n = 1558) and mH2A double KO (n = 1558) (two replicates each). c WNN UMAP plot showing three cell types (Basal, n = 1113; Luminal progenitors, n = 1891; Luminal Mature, n = 112) identified after clustering by graph-based clustering of the combined scRNA-seq and scATAC-seq signals and identifying the cell-types using markers. d Volcano plot showing differentially expressed genes in each cell type tested (separately) using Mann–Whitney U test with Bonferroni correction. e Violin plot showing the number of interactions per open chromatin region in wt (n = 199052) and mH2AdKO (n = 217727) with interactions (co-accessibility score > 0.1) predicted from the scATAC-seq data using Cicero. Mean and p-values shown were obtained by comparing means using Mann–Whitney U test. f Volcano plot showing the differential enrichment of ChIP-seq peaks of DNA binding proteins from ReMap in open chromatin regions in each cell type tested using Fisher’s exact test. Open chromatin regions per cell type were chosen as peaks that had a non-zero number of cut-sites in at least 10% of the cells of that cell type. g Heatmap showing the enrichment of gene expression of transcription factors in each cell-type (enrichment represented by −log10 (p-value) from Mann–Whitney U test with Bonferroni correction on gene expression). h Heatmap showing the motif enrichment of transcription factors in each cell-type (enrichment represented by -log10 (p-value) from Mann–Whitney U test with Bonferroni correction on chromVAR motif enrichment scores). LP luminal progenitors, ER estrogen, fMaSC fetal mammary stem cells, LM luminal mature.