Summary
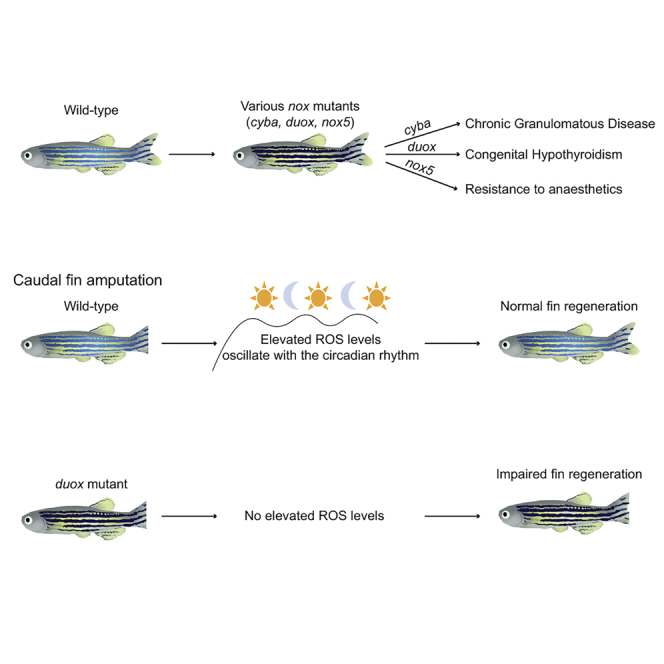
Sustained elevated levels of reactive oxygen species (ROS) have been shown to be essential for regeneration in many organisms. This has been shown primarily via the use of pharmacological inhibitors targeting the family of NADPH oxidases (NOXes). To identify the specific NOXes involved in ROS production during adult caudal fin regeneration in zebrafish, we generated nox mutants for duox, nox5 and cyba (a key subunit of NOXes 1–4) and crossed these lines with a transgenic line ubiquitously expressing HyPer, which permits the measurement of ROS levels. Homozygous duox mutants had the greatest effect on ROS levels and rate of fin regeneration among the single mutants. However, duox:cyba double mutants showed a greater effect on fin regeneration than the single duox mutants, suggesting that Nox1-4 also play a role during regeneration. This work also serendipitously found that ROS levels in amputated adult zebrafish fins oscillate with a circadian rhythm.
Subject areas: Cell biology, Ichthyology, Molecular genetics
Graphical abstract
Highlights
-
•
Fin amputation results in elevated ROS that oscillate with the circadian clock
-
•
duox mutants have diminished ROS levels following caudal fin amputation
-
•
duox mutants have diminished rate of caudal fin regeneration
-
•
duox/cyba mutants have a more severe defect in regeneration than duox mutants
Cell biology; Ichthyology; Molecular genetics
Introduction
A major aim in the field of regenerative biology is to uncover the mechanisms employed by various species in the animal kingdom, enabling them to regenerate organs and appendages, and in some cases, entire bodies. Deciphering such mechanisms, especially those that are evolutionarily conserved, will provide insight into why some animals have greater regenerative potential than others. Importantly, uncovering those highly conserved mechanisms may lead to novel therapies that could awaken a better regenerative response, clinically, in the sister field of regenerative medicine.
Over the past decade, it has become increasingly apparent that elevated reactive oxygen species (ROS) levels play an essential role during whole-body, appendage, organ, and tissue regeneration across the animal kingdom, including Hydra, planarians, Drosophila, zebrafish, axolotl, Xenopus, reptiles, and mammals.1,2,3,4,5,6,7,8,9,10,11,12 A key outstanding question is what are the mechanisms that control ROS production following injury and during regeneration? Findings from several model organisms have shown a critical role for Nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (NOXes) in the production of ROS following injury.5,7,8,9,10,11,13,14 The NOXes comprise a family of transmembrane, sometimes multi-component enzymes, whose primary physiological function is the transfer of an electron across biological membranes onto molecular oxygen to produce superoxide. The superoxide ion, which is an ROS, can then be dismutated to other more stable and readily diffusible ROS forms, such as hydrogen peroxide (H2O2).15 Most previous work establishing a critical role for NOXes during ROS production and regeneration has been based on the use of pharmacological inhibitors such as diphenylene iodonium (DPI), apocynin, and VAS2870, which lack specificity and selectivity.16,17 Therefore, to identify the specific NOXes responsible for ROS generation during adult appendage regeneration, we decided to take a genetic approach using zebrafish. All fins of the adult zebrafish are able to regenerate.18 We focused on the caudal fin, which is easily accessible and has unlimited regeneration potential.19 Here we report the generation and characterization of several mutant alleles of zebrafish, cyba, nox5, and duox. Using homozygous mutants for each of these genes, as well as cyba:duox double mutants, we investigated the effect of each mutant on ROS production and adult fin regeneration.
Results
Molecular characterization of nox mutant alleles
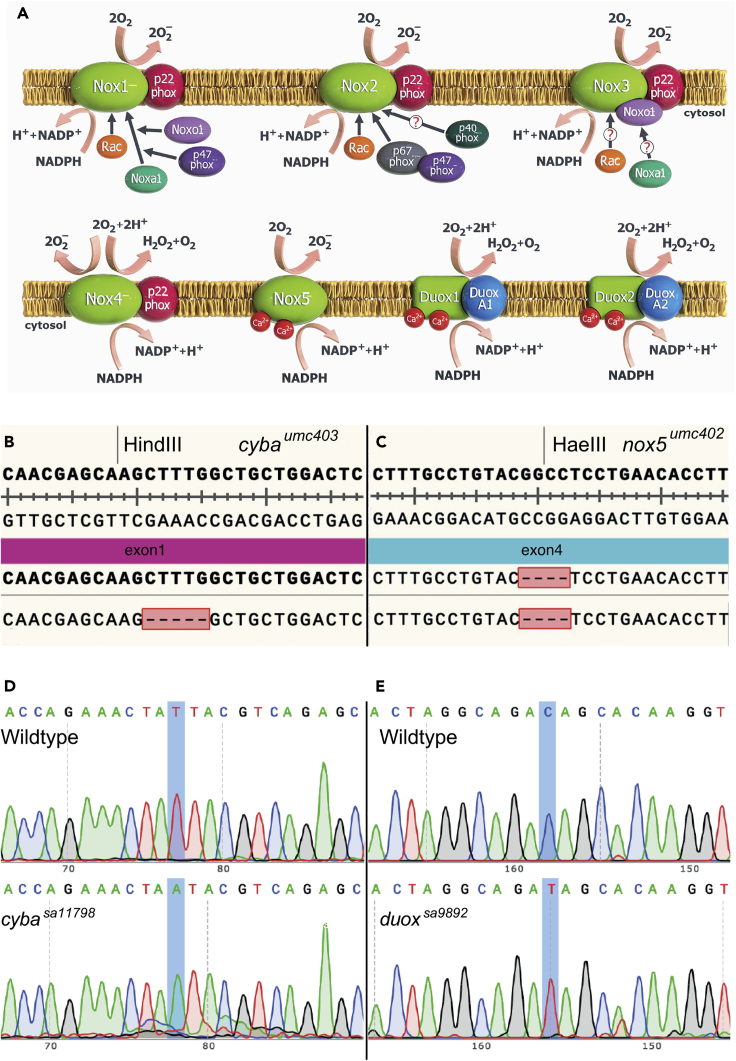
The primary aim of this work was to identify the molecular mechanisms responsible for sustained ROS production during zebrafish adult caudal fin regeneration. Previous work has provided evidence that NOXes play an essential role in ROS production and subsequent caudal fin regeneration, but that work was based on the use of NOX inhibitors,13 which inhibit all NOXes.16 Thus, we chose a genetic approach as a more specific strategy to pinpoint the NOXes. To do so, we sought to obtain or generate mutant alleles for a number of NOXes or their essential subunits (Figure 1A). One such critical subunit, P22phox, is encoded by the CYBA gene. P22phox plays an essential role in the maturation and structural integrity of NOXes 1–4.20 Like the mammalian CYBA orthologues, the zebrafish cyba genomic locus contains 6 exons and 5 introns.21,22 We also generated a mutant allele for nox5, which encodes Nox5, a calcium-regulated single-subunit NOX enzyme (Panday et al., 2015). NOX5 is present in humans but is absent in rodents.23 The zebrafish nox5 has 2 splice variants, both of which are protein coding and feature 16 exons. To try to achieve null mutant alleles for cyba and nox5, we targeted exon 1 of cyba and two sequences in exon 4 (common to both splice variants) for nox5 using CRISPR. F0 adults were crossed to wild-type (WT) animals, and F1 cybaumc403/+ and nox5umc402/+ animals were identified via sequencing and subsequently via restriction digest with HindIII and HaeIII, respectively (Figures 1B and 1C). For cyba, we found a 5bp deletion, leading to a frameshift mutation. For nox5, a 4bp indel resulted from the CRISPR-induced mutagenesis, also resulting in a frameshift mutation. Incrossing F1 heterozygotes led to the establishment of stable homozygous mutant lines, hereafter referred to as cybaumc403 and nox5umc402. For cyba, we also obtained a nonsense mutant allele, cybasa11798, from the Zebrafish Mutation Project.24 cybasa11798 is located in exon 4 (Genome assembly: GRCz11), leading to a T>A transversion (Figure 1D) causing a premature stop codon (TAG) after the 87th amino acid of the protein. Homozygous mutants for cyba and nox5 are viable and fertile and display no overt visible phenotypes. The zebrafish genome encodes a single duox gene, instead of the two paralogues (DUOX1 and DUOX2) present in tetrapods.25 We have previously characterized a duox nonsense mutant allele (duoxsa9892),26 which also arose from the Zebrafish Mutation Project.24 The duoxsa9892 mutation is located in exon 21 (Genome assembly: GRCz10), resulting in a C>T transition (Figure 1E) and a premature stop codon (TAG) after the 944th amino acid. Homozygous mutants for this allele are viable as adults and display congenital hypothyroidism.26
Figure 1.
The family of NADPH oxidases
(A–E) All NOXes are transmembrane proteins that transport oxygen across biological membranes, reducing oxygen to superoxide, which can then undergo dismutation to generate H2O2. NOXes are multi-componential proteins, and p22phox (the product of the cyba gene) is a common subunit to NOX1-4. Zebrafish lack Nox3 and have a single Duox isoform. Mutants for cyba (B) and nox5 (C) were generated using CRISPR, resulting in a 5bp deletion (cybaumc403) and a 4bp indel (nox5umc402), respectively. The mutations led to the removal of restriction enzyme sites, providing an easy diagnostic method for screening animals. Characterization of cybasa11798 (D) and duoxsa9892 (E) via Sanger sequencing shows the single nucleotide changes T>A and C>T, respectively, in contrast to a WT reference sequence. Panel A courtesy Dr. Kalin Narov, https://kalinnarov.wixsite.com/embryosafari.
See also Figures S1 and S2.
cyba and nox5 mutations are linked to unique phenotypes
After generating the cyba, duox, and nox5 mutant alleles, we asked whether any of them displayed homozygous mutant phenotypes. We recently showed that duoxsa9892 mutants are homozygous viable but display several phenotypes consistent with congenital hypothyroidism, including shorter body length, external goiters, hyperpigmentation, and sterility. These phenotypes could be rescued by T4 dosing and could be phenocopied in WT animals via chronic exposure to a goitrogen.26
For cyba, we took clues from its clinical relevance to chronic granulomatous disease (CGD). In human patients, CGD is often linked to mutations in CYBA.22 To this end, CGD patients experience recurrent infections, most frequently brought on by Aspergillus spp., Burkholderia cepacia, Staphylococcus aureus, Serratia marescens, Nocardia spp., and Salmonella.27,28,29,30 This high sensitivity to bacterial and fungal infections arises from inappropriate or insufficient ROS production in innate immune cells, required to kill engulfed pathogens. Although both zebrafish cyba mutant alleles used in this work are homozygous viable and fertile, we wondered whether they might be more susceptible to fungal infections and thus exhibit CGD-like phenotypes. Indeed, the cyba mutant line we obtained from the European Zebrafish Resource Center (EZRC) (cybasa11798) had been previously shown to be highly sensitive to fungal infections.31 However, we wished to confirm whether the CRISPR-mediated mutant we generated, cybaumc403, was also hypersensitive to fungal infections. Given that invasive infections by filamentous fungi are especially important contributors to morbidity and mortality in CGD patients,32,33,34 we asked whether cybaumc403 larvae had increased susceptibility to Aspergillus fumigatus infections, relative to WT larvae. To achieve this, we took advantage of the large and easily accessible larval yolk sac to inject a suspension of A. fumigatus conidia into cyba mutant and WT larvae. We first asked whether injecting buffer alone could bring about mortality. For this we injected buffer solution in both cyba mutant (n = 50) and WT larvae (n = 50) and compared the mortality rates in those versus uninjected cohorts (n = 50 each). The larvae of both genotypes tolerated the injections well, with fewer than 3 deaths each after injection, irrespective of genotype. However, across two independent experiments (Figures S1A and S1B), cybaumc403 animals (n = 25) injected with A. fumigatus conidia showed a significantly higher mortality rate (40% in A and 86% in B) at 3dpi, compared to WT larvae (n = 17) (17% in A and 27% in B) (log rank test; p value, <0.005) (Figures S1A and S1B). Similar results were obtained following injection of A. fumigatus conidia into cybasa11798 larvae (n = 8), with mortality rates at 87% in this instance (Figure S1A). All larvae that developed infections showed similar phenotypes. During the initial stages of infection, the swimming ability of the larvae was significantly affected whereby they could move their pectoral fins but could not achieve productive movement. As the infection advanced, larvae could not remain upright, probably due to effects on the swim bladder (Figures S1C and S1D). Terminal infections were characterized by extensive necroses and feebly moving gill arches and heartbeat. Thus, in agreement with the previous study,31 cybasa11798 animals were very sensitive to A. fumigatus infection. Similarly, cybaumc403 animals were also ultrasensitive to A. fumigatus infection. This confirmed that mutations in cyba cause high susceptibility to fungal infection in zebrafish larvae, as in human patients with CGD, thus designating both cyba mutant alleles as useful models for human CGD.
Based on the existing literature, we had no clues as to the phenotypes that we might expect in homozygous nox5 mutant fish. No human disease has yet been associated with NOX5 mutations, and rodents lack a Nox5 ortholog. While we found nox5ex4.4bp fish to be viable and fertile, we serendipitously uncovered an unusual mild phenotype in these animals after treating them with anesthetics. Intriguingly, most nox5ex4.4bp animals appeared to have prolonged resistance to the commonly used anesthetic, tricaine methanesulfonate (MS-222). To characterize this phenotype further, we individually treated nox5ex4.4bp adults (n = 22) and WT adults (n = 23) with MS-222 (0.04%) in a beaker. We then measured two responses to the anesthetic: 1) righting reflex and 2) cessation of opercular movement. The righting reflex normally serves to resume orientation when the body loses its upright orientation. All WT animals lost the righting reflex in under 30 s following immersion (Figure S2A). After this point, simple tapping of the bench on which the beaker was placed elicited no response from them. In contrast, it took nox5ex4.4bp animals up to 360 s to achieve loss of the righting reflex. Even though they stopped swimming, mutants remained responsive to the tapping, which stimulated jerky, erratic movements. The loss of opercular movement in WT animals took up to 200 s, while in the mutants this took as long as 382 s (Figure S2B). Interestingly, most mutants appeared to lose the righting reflex very close to cessation of opercular movement. We further checked the response of both groups to lidocaine hydrochloride and cold water, both of which have been previously tested as anesthetics in zebrafish.35 While neither group achieved a surgical plane of anesthesia with 750 mg/L of lidocaine, loss of the righting reflex was significantly delayed among the nox5 mutants (Figure S2A). Meanwhile, cold water induced a rapid surgical plane of anesthesia on both groups, with no statistically significant difference in their responses (Figure S2).
Overall, we found that, while all nox mutant alleles were homozygous viable into adulthood, each displayed unique phenotypes, suggesting that all mutants were at least hypomorphic, if not null mutant alleles.
Caudal fin regeneration in zebrafish is not influenced by age
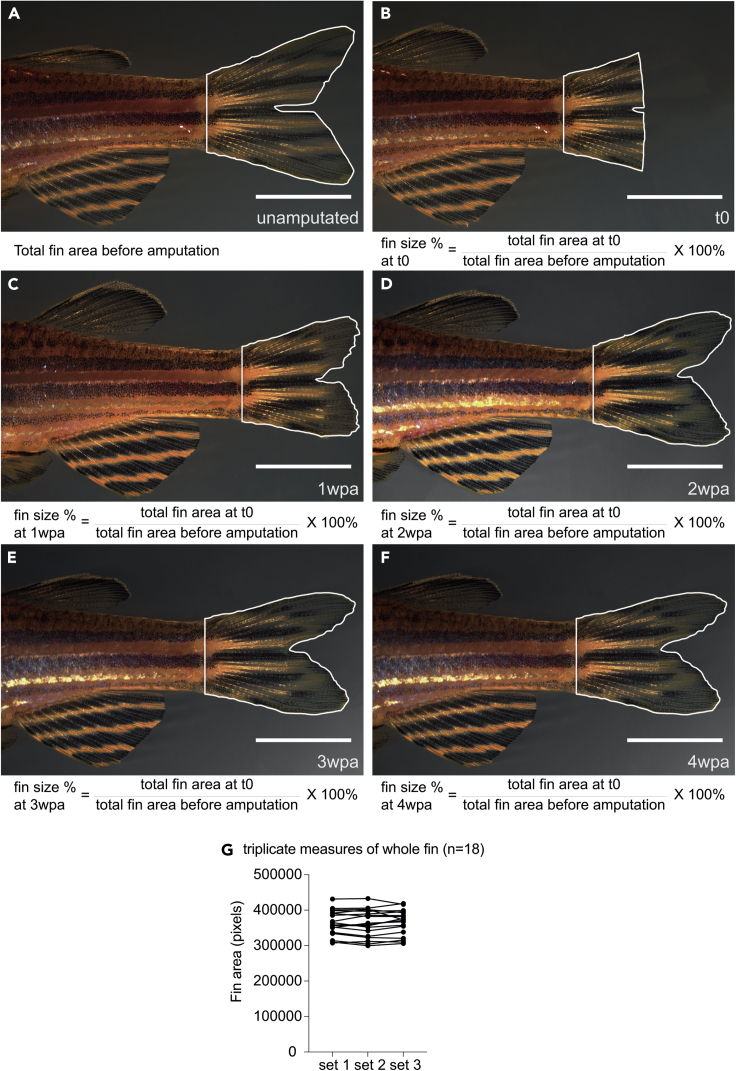
Given that we would be assessing fin regeneration in animals of different genotypes and ages throughout this study, we performed caudal fin amputations in WT fish of different ages to ascertain whether their age might affect the extent and/or rate of fin regeneration. For this analysis, we amputated approximately 50% of the caudal fin in adult animals from 3 months through 22 months of age and then followed them individually over the four weeks it normally takes for fin regeneration to complete. Given that we were interested in regeneration of the entire fin, as opposed to specific parts of the fins, such as the rays, we first measured the size of the entire caudal fin of individual animals immediately prior to and after amputation. We then measured the caudal fins of each individual once a week for up to four weeks and determined the extent of regeneration as shown in Figures 2A–2F. However, we were cognizant of the potential variability in measuring the fin area, due to the variable extent of fanning out of the fin during repeat measurements over time. To investigate whether this might be an issue, we first performed repeat measurements on 18 individual animals prior to amputation (where each individual was anesthetized, imaged, and allowed to recover for a total of three times). We found that, if the animals are allowed to relax fully following each anesthesia event before imaging, the fin area measurements were very similar over the triplicate measurements (Figure 2G). The variability across the triplicate measurements per animal was generally between 1% and 4%, and there was no statistical difference between the triplicate measurements within individuals (Table S1). In contrast, there was a significant difference in the fin areas between individuals (Table S2), which is unsurprising given that zebrafish display markedly different growth rates even from the same clutch.36 Thus, given the greater variability between animals than within repeat measurements within individuals, we decided to assess regeneration by following individuals, rather than assessing regeneration by following the population, as is routinely done in the field.37,38,39 Using this approach, we measured the extent of regeneration in individual animals of different ages and we found that adult zebrafish younger than six months of age generally overshot the initial size of their fins over the four weeks after amputation (i.e., regenerated to over 100% of their initial fin size) (Figures S3A–S3C). However, caudal fins in fish six months and older generally only grew back to their original fin size (Figures S3D–S3I). From this we can conclude that caudal fin regeneration in young adult fish results from a combination of fin regeneration and growth, while in fish older than 6 months regeneration of the fin is not confounded significantly by fin growth as long as regeneration is not followed over more than four weeks post-amputation. We thus decided to concentrate the remainder of our tail fin amputation experiments on fish six months or older.
Figure 2.
Methodology for quantifying rate of adult fin regeneration
(A–F) Amputated adult caudal fins in zebrafish achieve complete, scar-free regeneration by 4 weeks post-amputation (wpa). In all cases, the fin was imaged prior to amputation (A), immediately following amputation (B) and then once weekly at 1wpa (C), 2wpa (D), 3wpa (E), and 4wpa (F). The distinctive stripe and pigment patterns on the body and anal fin enabled identification of individual animals. Fins were amputated midway, and the caudal peduncle was used to demarcate the proximal extent. Scale bar = 5 mm. (G) Repeat (triplicate) measurements of caudal fin areas in 18 adult fish (n = 18).
See also Figures S3, S4, Tables S1, and S2.
Next, we were interested in addressing whether the rate of fin regeneration is affected by the age of the fish. To answer this question, we plotted the slope of regeneration over the first, second, third, and fourth week of regeneration as a function of age (Figures S4A–S4E). We found that, overall, caudal fin regeneration is the fastest during the first two weeks of regeneration (week 0–1 and week 1–2) and then slows down during the third (week 2–3) and fourth week (week 3–4) (Figure S4F). Thus, the rate of caudal fin regeneration is biphasic, with similar rates between the first and second weeks and between the third and fourth weeks, with the latter two statistically slower than the first two weeks (Figure S4F). Furthermore, we found that animals between six and eight months of age regenerated at the same rate over the four-week period, while older animals exhibited statistically different regeneration rates relative to animals of other ages during at least one time point (Figures S4A–S4E). These results provided us with a good “working age range” of animals, where neither growth nor rates of regeneration were significantly affected by the age of the animals, and thus we concentrated subsequent analyses on animals between six and eight months of age.
cyba and duox mutations affect caudal fin regeneration
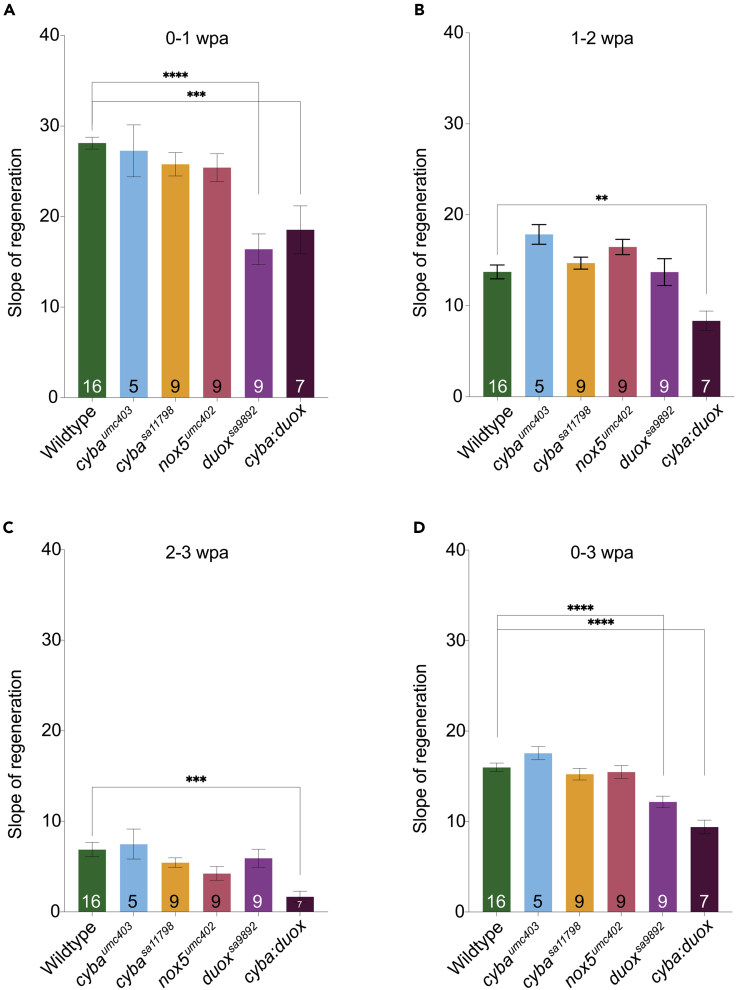
Having identified regeneration trends in WT animals, we proceeded to investigate the rate of regeneration in the various nox mutants, namely cybaumc403 (n = 5), cybasa11798 (n = 9), duoxsa9892 (n = 15), nox5umc402 (n = 9), and cybasa11798:duoxsa9892 double mutants (n = 7) (hereafter referred to as cyba:duox), along with their WT siblings (n = 16) (Figures 3 and S5). All caudal fin amputations were performed on animals between six and eight months of age. In addition, we took care to include only animals that had similar amounts of the caudal fins amputated (between 40% of 55% of the original fin size) for subsequent analyses as evidence from others has shown that the rate of fin regeneration may be affected by the amount of fin that is amputated.39,40,41 Once all of these factors were taken into consideration, we found that, out of the various nox mutants, only duoxsa9892 and cyba:duox animals displayed a significant effect on the rate or extent of adult caudal fin regeneration (Figure 3). More specifically, we found that duoxsa9892 animals had a significantly slower rate of regeneration when compared to WTs during the first week following amputation (Figure 3A). This delayed rate of regeneration was further amplified in the cyba:duox double mutants, where the slower rate of regeneration continued into the third week (Figures 3B and 3C). It is important to point out that, while regeneration in these two mutant strains was not entirely inhibited, these mutants exhibited a significant slowing in the rate of regeneration over the 3 weeks that they were allowed to regenerate. In particular, the overall size of the regenerating fins in duoxsa9892 and cyba:duox animals remained significantly smaller than that in the WT cohorts (Figure S6). Thus, the overall defect in regeneration appeared to be in the rate of regeneration, more so than on overall regeneration per se. To identify the predicted timescales expected for the completion of regeneration, i.e., the time likely to take for the fins to reach the unamputated size, we used row statistics with slopes derived from simple linear regression (Figures S7 and S8). This revealed that WTs, cyba, and nox5 mutants would complete regeneration in under 5 weeks post amputation (wpa) (Figures S8A–S8D), while duoxsa9892 animals would not be expected to reach their pre-amputation sizes until 7.3wpa (Figure S8E), and cyba:duox animals would not achieve their pre-amputation sizes until 11.2wpa (Figure S8F). Thus the duoxsa9892 and cyba:duox mutants displayed a 2–3 times slower rate in tail fin regeneration relative to WT animals or the other nox single mutants assessed in this study.
Figure 3.
duox mutants regenerate the caudal fin slower than WTs
(A–C) Significant differences in regeneration were observed between WT and duox mutants during 0-1wpa (A) and 1-2wpa (B). By 2-3wpa (C) these differences were resolved, with only the double mutant cyba:duox continuing to significantly trail behind.
(D) An overall view across 0-3wpa highlights how duoxsa9892 and cyba:duox animals are significantly slower than every other group. Asterisks denote statistically significant differences (Bonferroni’s multiple comparisons test, ∗p < 0.5, ∗∗p < 0.01, ∗∗∗∗p < 0.0001). WT (n = 16); cybaumc403 (n = 5); cybasa11798 (n = 9); nox5umc402 (n = 9); duoxsa9892 (n = 9); cyba:duox (n = 7).
See also Figures S5–S8.
The effect of hypothyroidism on the rate of fin regeneration
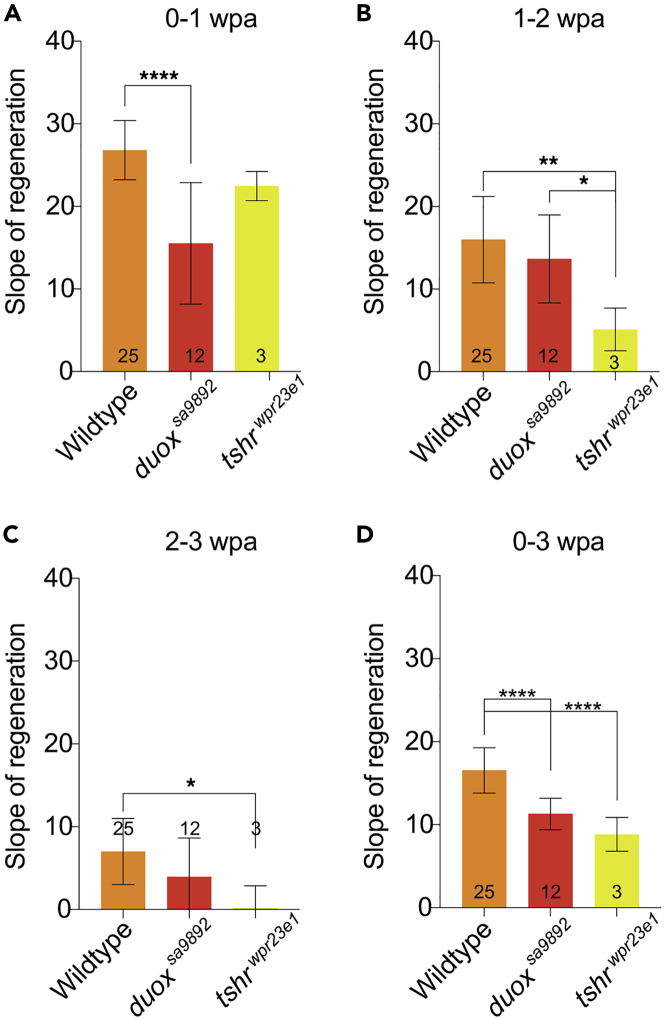
Recently, we reported that duoxsa9892 mutant zebrafish exhibit congenital hypothyroidism.26 We thus wondered whether the reduced rate of regeneration among the duoxsa9892 animals could be attributed to their hypothyroidism. We therefore investigated fin regeneration using another zebrafish model of congenital hypothyroidism, namely the tshrwpr23e1 mutant (original name manetwpr23e1).42 This mutant harbors a nonsense mutation in exon 2 in the tshr gene, which encodes the thyroid-stimulating hormone (TSH) receptor. We have previously reported phenotypic concordance between tshrwpr23e1 and duox mutants.26 Conveniently, the tshrwpr23e1 allele destroys an HpyCH4V restriction site, providing an easy diagnostic method for genotyping. The effect of the tshr mutation on body length had not been previously reported, so we measured the body length of WT, tshrwpr23e1/+, and tshrwpr23e1 siblings at 3 months of age. This assessment showed a similar growth retardation phenotype that we previously reported in duoxsa9892 mutants26 (Figure S9). We proceeded to amputate the caudal fin in WT, tshrwpr23e1, and duoxsa9892 fish. In the first wpa, a significantly slower rate of regeneration was observed in duoxsa9892, when compared to WT animals or tshrwpr23e1 fish (Figure 4A). However, during the second and third weeks of regeneration, the duoxsa9892 and WT fish had no significant difference in their rates of regeneration (Figures 4B and 4C). In contrast, the tshrwpr23e1 fish regenerated at the same rate as WT fish during the first week (Figure 4A), but then their regeneration rate slowed down relative to both WT and duoxsa9892 fish during the second and third weeks of regeneration (Figures 4B and 4C). In summary, both duoxsa9892 and tshrwpr23e1 animals displayed delayed rates of fin regeneration over the three-week period (Figure 4D). However, the duoxsa9892 animals displayed the greatest impact on fin regeneration during the initial phases of regeneration, while hypothyroidism alone decelerates it only during the later stages, suggesting that the effect of duox on regeneration is not solely due to their hypothyroidism.
Figure 4.
Thyroid hormone deficiency slows down caudal fin regeneration but only during the late phases of regeneration
(A–C) The impact of duoxsa9892, but not tshrwpr23e1, on the rate of regeneration is significant during 0-1wpa. By 2wpa (B) the effect of tshrwpr23e1 becomes significant, and this effect continues into 2-3wpa (C).
(D) An overall view across 0-3wpa indicates duoxsa9892 and tshrwpr23e1 animals are significantly slower than WTs during regeneration. Asterisks denote statistically significant differences (Bonferroni’s multiple comparisons test, ∗p < 0.5, ∗∗p < 0.01, ∗∗∗∗p < 0.0001). WT (n = 25); duoxsa9892 (n = 12); tshrwpr23e1 (n = 3).
See also Figure S9.
ubb:HyPer is an effective reporter of amputation-induced ROS production
Given that the slower rate of regeneration during the first week in the duox mutants is not linked to their hypothyroidism, we then asked whether it might be due to a diminished production of ROS in the mutants during the early phase of regeneration. To test this hypothesis, we generated a transgenic line that expresses the H2O2-specific ROS sensor, HyPer,43 under the control of the ubiquitous ubb promoter (ubb:HyPer), referred to as Tg(ubb:HyPer)umc400. We first measured ROS levels in unamputated caudal fins of individual control Tg(ubb:HyPer)umc400 adults (n = 7) (Figures S10A and S10B), following each animal over the course of four days. Individual animals were identified based on their scale morphology, which is very distinctive under fluorescence. Using a set of repeated time points during the day over four days, we aimed to identify the “baseline” H2O2 levels in adult fins. While basal H2O2 levels in uninjured fins were relatively stable throughout the four-day period (Figure S7B), ROS levels rose significantly above this baseline level following fin amputation (Figures S10C and S10D). For consistency, we performed all fin amputations around midday. Observations were recorded at the same time intervals as those used for the unamputated controls, with additional time points at 2, 6, and 10 hours post amputation (hpa) (Figure S10D). Unlike the unamputated cohort, these were followed for two weeks. By 2hpa (around 3 p.m.), ROS levels showed a significant increase (paired t-test, p = 0.0009) above the unamputated level measured at the same time. At 6hpa and 10hpa, the increase in ROS levels was sustained, but thereafter the levels appeared to oscillate each day with the highest levels in the mornings (between 7 a.m. and 8 a.m.) and the lowest levels in the afternoons (between 3 p.m. and 4 p.m.) (Figures 5A and S10D). This rise and fall in ROS levels was recorded over three consecutive days. After three days, the frequency of recordings was reduced to once daily, between 3 p.m. and 4 p.m. After the sixth day, the frequency of imaging sessions was further reduced to once every two days, between 3 p.m. and 4 p.m., at which point ROS levels were still higher than the baseline, pre-amputation levels (Figure S10D). The final recording was made mid-afternoon (between 3 p.m. and 4 p.m.) two weeks after amputation. At this point the ROS levels had finally returned to pre-amputation baseline levels (Figure S10D).
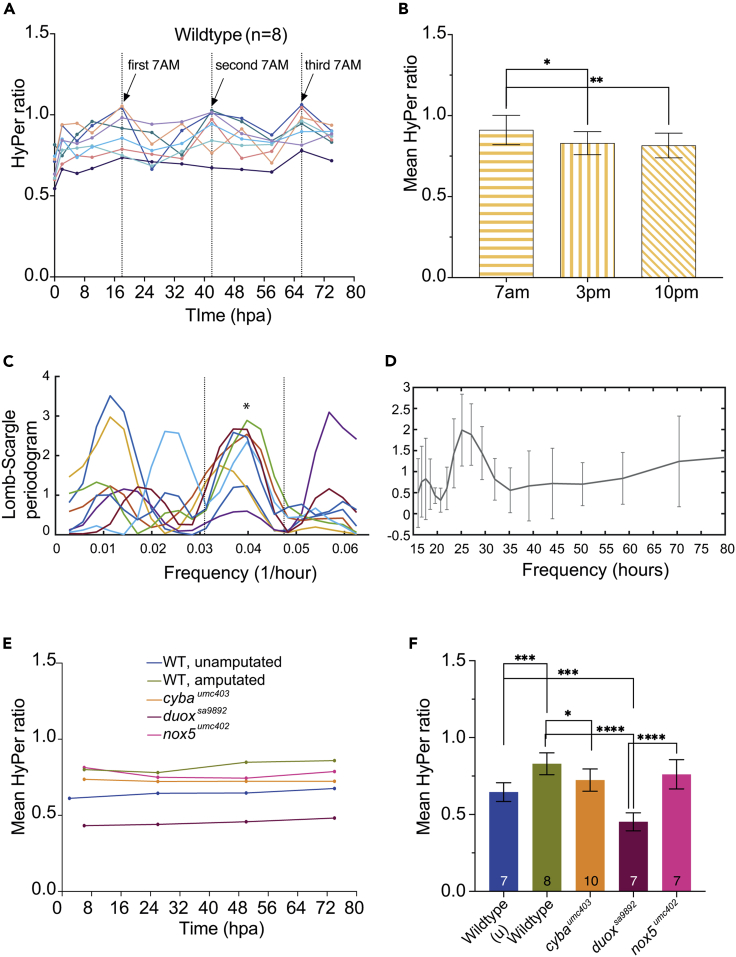
Figure 5.
HyPer ratios in WT animals and nox mutants
WT animals and nox mutants were generated in a Tg(ubb:HyPer)umc400 transgenic background, which permits in vivo assessment of ROS levels. (A) ROS Hyper ratios in individual WT animals (n = 8) observed at three time points during the day over three days post-amputation reveal an oscillatory pattern of ROS levels.
(B) ROS levels over three consecutive days post-amputation were significantly higher in the mornings (7a.m.) than in the afternoons (3 p.m.) or nights (10p.m.).
(C and D) Long-Scargle periodogram reveals that the ROS oscillations follow a daytime-dependent trend, with a periodicity of around 0.4 (∗ on graph), indicative of a ∼24 h circadian cycle (D).
(E) Graph of ROS levels measured at the same time of day (3p.m.) in WT (unamputated), WT (amputated), cybaumc403, duoxsa9892, and nox5umc402 animals.
(F) Graph of mean Hyper ratios measured over three consecutive days at 3p.m. reveals significantly attenuated post-amputation ROS levels among the duoxsa9892 mutants relative to the WT amputated fins and the other nox mutants. Indeed, ROS levels in the duoxsa9892 fins are significantly lower than even those found in unamputated WT fins. ROS levels in the amputated cyba mutant fins are significantly lower than those present in WT amputated fins but not as low as in the duoxsa9892 fins. ROS levels in the amputated nox5umc402 mutant fins are not significantly different from those present in amputated WT fins. (Bonferroni’s multiple comparisons test, ∗p < 0.5, ∗∗p < 0.01, ∗∗∗∗p < 0.0001). For A-D, WT (n = 8). For E-F, WT unamputated (n = 7); WT amputated (n = 8); cybaumc403 (n = 10); duoxsa9892 (n = 7); nox5umc402 (n = 7).
See also Figures S10 and S11.
We then asked whether the ROS levels measured over consecutive days taken at the same time of the day differed significantly from ROS levels measured at other times of the day. This analysis showed that ROS levels measured at 7 a.m. were significantly higher than those measured at either 3 p.m. or 10 p.m., over the three consecutive days (Figure 5B). To confirm the veracity of these apparent oscillations, we then performed a Fourier transform power spectrum analysis on the changing ROS levels of all the amputated fins over the three days of measurements (Figures 5C and 5D). This analysis showed a coordinated high periodicity peak in ROS levels in all fish, at a frequency of 0.04 1/h (equating to a periodicity of ∼24 h) (Figures 5C and 5D). Thus, we found that adult fin amputation led to a sustained increase in ROS levels over the first two weeks of regeneration and that ROS levels in the amputated fins oscillate with a circadian rhythm.
Post-amputation ROS production is affected in the nox mutants
Having determined the post-amputation sustained increase in ROS levels in WT fish (Figures S10D and S11A), we next crossed or generated genetically altered lines for all the nox mutants in the Tg(ubb:HyPer)umc400 background. We then assessed whether post-amputation ROS levels were altered in these mutants. We found that the cybaumc403 animals (n = 10) displayed an increase in ROS levels post-amputation (t-test, p = 0.0004) (Figure S11B). Interestingly, the cyba mutants returned to pre-amputation ROS levels by 1wpa (168hpa) (Figure S11B), in contrast to the WT animals, where pre-amputation ROS levels were not seen until 2wpa (Figure S10D). Also, the oscillations in ROS levels during the day exhibited peaks of smaller amplitudes and had a lower H2O2 increase overall when compared to WT fish. Furthermore, H2O2 levels in cyba mutants were similar to those found in WT fish at 2hpa but had lower values at later time points.
In contrast, duoxsa9892 animals (n = 7) (Figure S11C) showed low ROS levels post-amputation throughout the week-long course of observations. The post-amputation levels were significantly lower at all time points taken, and there were no distinctive peaks associated with the 7 a.m. time points. Even more interestingly, ROS levels were lower than those present in WT unamputated controls (Figures 5D and S11A and S11C). Finally, the nox5ex4.4bp animals (n = 8) displayed a significant increase in ROS levels at 2hpa (3 p.m.) (t-test, p = 0.04), which peaked at 7 a.m. (18hpa) the next morning (t-test, p = 0.036), and rapidly “baselined”, showing no further significant increase thereafter (Figure S11D). In order to better compare the HyPer ratios between the different genetic backgrounds we superimposed the ratios obtained for all the genetic strains in Figure S11E. Given that we previously found significant differences between the HyPer ratios in WT animals post-amputation depending on the time of the day, we also compared the HyPer ratios across all the nox homozygous mutant strains observed at the same time of day post-amputation (i.e. 3 p.m.) over the first three days post-amputation alongside the ratios found in unamputated tail fins (Figure 5D). This analysis confirmed that duoxsa9892 animals exhibited the most profound effect on ROS levels following amputation, where no increase was observed in these animals. Furthermore, it confirmed that duoxsa9892 tail fins have significantly lower ROS levels even in the pre-amputation state, when compared to all other genotypes. The other nox mutants had a much milder effect on post-amputation ROS levels, and unlike the duox mutants, none of the other nox mutants affected the initial increase in ROS levels caused soon after fin amputation (Figure 5D).
Overall, these results pinpoint Duox as the primary Nox responsible for ROS production following injury and during regeneration, with the other NOXes also potentially involved in ROS production but to a much lesser extent. In particular, cyba mutants returned to pre-amputation ROS levels after 1wpa, in contrast to WT animals, where elevated ROS levels were sustained over the first 2wpa. This might help explain why the cyba:duox double mutants have a stronger effect on regenerative capacity than the duox single mutants during the later phases of tail fin regeneration.
Discussion
In this work, we set out to uncover the mechanisms of ROS production following zebrafish caudal fin amputation, with particular emphasis on the NOX family of enzymes. While an essential role for ROS production, mediated primarily by NOXes, in whole organism and appendage regeneration is well documented in the literature, most studies addressing this role were primarily based on the use of chemical inhibitors targeting unspecifically the entire family of NOXes.2,5,7,8,9,10,11 In an attempt to pinpoint which NOXes are primarily responsible for ROS production following injury and subsequent regeneration in adult zebrafish, we employed a genetic approach in this study. An initial concern arising from this was the potential complication of genetic redundancy present in teleost fish. Following the divergence of the ray-finned and lobe-finned fishes, teleosts underwent an additional whole genome duplication event in the common ancestor of zebrafish and the other 22,000 ray-finned species.44 Despite this concern we found that the zebrafish genome does not harbor additional duplicated paralogues for cyba, duox, or nox5 when compared to other non-teleost vertebrates. In fact, we and others have found that zebrafish has only a single duox gene, as opposed to most other vertebrates, including mice and humans, which have two duox paralogues (DUOX1 and DUOX2).25,26 It should be noted at this point that the zebrafish genome currently has a gene mis-annotated as duox2 (ZBD GENE 111007-1). This gene is not a paralog of duox but instead is the zebrafish paralog of duoxa, which encodes a Duox maturation factor.45 Duox maturation factors are ER proteins that are essential partners of the DUOXes, helping prevent their retention in the ER.15 In conclusion, the zebrafish genome contains only a single duox paralog, as opposed to most other vertebrates, which harbor two paralogues. Therefore, our work suffered less complications arising from excessive genetic redundancy than it would have if it were carried out in most other vertebrate model systems.
Another potential complication could have arisen if homozygous mutants for any of the genes used in this work had been non-viable as adults. Fortunately, this was not the case. Although viable, we found that homozygous mutants for all three genes did display phenotypes, ranging from hypothyroidism and sterility (duox) to enhanced susceptibility to fungal infections (cyba) and increased resistance to some anesthetics (nox5). Unsurprisingly, the phenotypes associated with duox and cyba mutations in zebrafish were consistent with mutations in the human orthologs of these genes, with DUOX2 and CYBA, leading to congenital hypothyroidism and chronic granulomatous disease, respectively.22,46 Thus, mutations in the zebrafish orthologs of these two genes provide useful models for these human genetic diseases. In contrast, there are currently no human genetic diseases associated with mutations in NOX5, and so we had no prior knowledge of what phenotypes might be associated with mutations in zebrafish nox5. In addition, no information can be inferred from mouse knockout studies as rodents lack a Nox5 orthologue. Serendipitously, we noticed nox5umc402 fish were particularly resistant to some anesthetics, including tricaine and lidocaine. However, we do not currently know the reason behind this. In summary, all the nox mutants we used in this work were homozygous viable and each presented with identifiable phenotypes, when appropriately challenged.
The influence of advancing age on regenerative decline is known among adult fish.47,48 Given that we would be using adults, we wanted to identify an age range where the rate of fin regeneration would not be affected by the age of the fish being used. We therefore performed fin regeneration assays, including rate of regeneration, in an array of age groups and found that young adults, from six to eight months of age, showed no significant difference in their rates of fin regeneration. We then set out to assess how each mutation, singly or in combination, affected ROS production following fin amputation and during fin regeneration. Our results showed that, among the single mutants, the duox mutants were the most striking, resulting in a significantly lower ROS levels and slower rate of regeneration, especially over the first two weeks following amputation. Furthermore, combinatorial mutants of duox and cyba displayed an even stronger effect on the rate of fin regeneration, extending into the third week following amputation, suggesting that Duox functions in combination with other NOXes during the later stages of adult fin regeneration in zebrafish. Further support that Duox works in combination with other NOXes to generate the full complement of ROS activated during caudal fin regeneration can be gleaned by the observation that addition of the chemical inhibitors targeting all NOXes resulted in a more pronounced diminution of ROS levels and a stronger effect on caudal fin regeneration13 than what we observed in this study in duox or duox:cyba mutant animals. Previously, genetic approaches have also identified a primary role for the Duox maturation factor in ROS generation during Drosophila imaginal wing disc regeneration,4 suggesting this role for Duox in regeneration may be an ancient, conserved function. This is likely due to the fact that Duox is regulated by calcium and a rise in intracellular calcium is a conserved rapid response following injury.14,49,50 Furthermore, previous studies have implicated NOX2 during axonal regeneration in mammals.12 Intriguingly, Nox2 is one of the multi-component NOXes, whose activity is dependent on Cyba, which encodes p22phox, also sometimes referred to as the α subunit of Nox2.20 Thus, our cyba zebrafish mutants would be expected to have impaired Nox2 activity. While we did not find an overt impairment in the rate of caudal fin regeneration in the cyba mutants, we found that these animals failed to sustain elevated levels of ROS beyond the first week post-amputation, unlike the WT controls. While this difference in ROS levels was not sufficient to result in a change in the rate of regeneration in the cyba alone mutants, we did find that the duox:cyba double mutants had a more pronounced effect on the rate of caudal fin regeneration, especially after the second week of regeneration, than duox alone mutants. Given that our work was primarily interested in overt appendage regeneration, we did not investigate whether the cyba mutants exhibited defects in axonal regeneration during caudal fin regeneration, but this would be an interesting question to explore in the future.
While the focal point of this work was the genetic dissection of the role of NOXes in ROS production during adult fin regeneration, our work uncovered some unexpected findings. One was that ROS levels not only rise after fin amputation but also oscillate with a circadian rhythm in the days following injury, with the highest levels found in the mornings (around 7 p.m.) and the lowest levels in the mid-afternoons (around 3 p.m.). Discovering this was important as it called for the need to measure ROS levels at the same time daily, for a more accurate assessment of post-amputation ROS levels in the WT versus mutant fish, without being complicated by the circadian-associated oscillatory changes at different times of the day. However, this finding raises several questions. For example, what is responsible for the circadian-associated oscillations in injury-induced ROS levels? One possible link might be the well-appreciated oscillations in metabolism that are known to be linked to the circadian clock.51 We noted that these oscillations were mostly apparent following injury, which might be linked to the injury-induced changes in metabolism that have previously been highlighted in both Xenopus tail regeneration and zebrafish appendage and heart regeneration.52,53,54,55,56 It is thus possible that the oscillating levels of ROS are direct or indirect outputs of the metabolic state in the fin following injury. It is also notable that wound healing in mice and humans is affected by the time of day that the injuries are incurred or if the circadian clock is disturbed.57,58,59,60,61 It will be interesting to investigate whether the time of fin amputation affects wound healing or the speed or quality of regeneration in zebrafish, and if so, whether these effects are linked to the dynamics or peak levels of ROS production following injury.
Limitations of the study
In this study, we used single and compound Nox mutants to assess the specific and combinatorial roles for different Nox during caudal fin regeneration. However, given that there are seven NOXes, it was too onerous a task to generate mutations targeting each of the seven NOXes, individually. Therefore, we chose to target the multi-subunit sub-family of NOXes, namely Nox1-4, by targeting their shared subunit, namely cyba. Thus, this study is unable to assess the involvement of Nox1 vs Nox2 vs Nox3 vs Nox4 during caudal fin regeneration but instead used an approach that would affect all of these at the same time. However, given that the cyba mutants had no discernible effect on caudal fin regeneration on their own and an effect is only found when these are combined with duox homozygous mutants, we believe that not having targeted each Nox separately is less of a concern. Secondly, it is not known whether the mutant alleles used in this study are null mutants or hypomorphs, and this would impact on the interpretation of the study. Furthermore, it is not known whether compensatory mechanisms, as described by El-Brolosy et al,62 may have led to less severe consequences of the mutations on ROS production and/or fin regeneration, given that all of these mutant alleles include either frameshift mutations or nonsense mutations and thus would be expected to stimulate nonsense-mediated mRNA decay. Finally, all the cells in the animals used in this study were mutant, and thus, it is difficult to know whether some of the effects on regeneration might have been caused, at least in part, by systemic effects of the mutations on fin regeneration, rather than local requirements for those genes on these processes; this is especially the case with the duox mutants, which we know can have systemic effects, including hypothyroidism. Another potential limitation in the interpretations of this study is that the measurements of ROS levels were performed using a genetic sensor, HyPer, which is sensitive to changes in ROS levels but is also sensitive to changes in pH levels. We could determine whether the changes in HyPer ratio are caused by changes in pH levels. For this we would have to establish a transgenic line that expresses a pH-sensitive, but ROS-insensitive, variant of HyPer, called SypHer,63 and cross this new line with all the nox mutants used in this work, finally followed by incrossing these into homozygosity. However, such a task would be very time consuming and laborious. Thus, we cannot entirely exclude the possibility that some of the changes in HyPer ratio we have documented are not solely due to changes in ROS levels or pH levels or a combination of both. However, we note that we previously addressed this issue during Xenopus tadpole tail regeneration, where we were able to show that tadpole tail regeneration is not associated with significant changes in pH levels, and thus changes in HyPer ratios during tadpole tail regeneration were entirely due to changes in ROS levels.5
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| ExTaq DNA Polymerase | Takara | RR001A |
| Proteinase K | New England Biolabs® | P8107S |
| Cas9-NLS protein | New England Biolabs® | M0646T |
| Experimental models: Organisms/strains | ||
| cybasa11798 | European Zebrafish Resource Centre | ZFIN ID: cybasa11798 |
| duoxsa9892 | European Zebrafish Resource Centre | ZFIN ID: duoxsa9892 |
| tshrwpr23e1 | David Parichy Laboratory | ZFIN ID: tshrwpr23e1 |
| Tg(ubb:HyPer)umc400 | This publication | ZFIN ID: Tg(ubb:HyPer)umc400 |
| nox5umc402 | This publication | ZFIN ID: nox5umc402 |
| cybaumc403 | This publication | ZFIN ID: cybaumc403 |
| Oligonucleotides | ||
|
cyba ex1.5bp F: 5’-AGTTTATTTGCCAGTGACAGCA-3’ |
Sigma-Aldrich® | Custom |
|
cyba ex1.5bp R: 5’-CTCAAGCAGCCTACCAAACC-3’ |
Sigma-Aldrich® | Custom |
|
cyba sa11798 F: 5’- CTGCTGGTGTGTTTGTGTGT -3’ |
Sigma-Aldrich® | Custom |
|
cyba sa11798 R: 3’- AAGGGACATGCTTGACAAGG -5’ |
Sigma-Aldrich® | Custom |
|
duox sa9892 F: 5’-ACGAGGTACACAACTCAAGCTG-3’ |
Sigma-Aldrich® | Custom |
|
duox sa9892 R: 5’-GACGTTCAAAGCGAAACCTGAC-3’ |
Sigma-Aldrich® | Custom |
|
duox sa9892 seq: 5’-CTTGGTCTGCCTTTGACGAAGT-3’ |
Sigma-Aldrich® | Custom |
|
manet F: 5’-TGCAAATTTCGATAAATTGTAATAA-3’ |
Sigma-Aldrich® | Custom |
|
manet R: 5’-GGTGAGGCTGCTTCATTTTC-3’ |
Sigma-Aldrich® | Custom |
|
nox5 ex4.4bp F: 5’-GCTCAAGGGCTTACATGATCC-3’ |
Sigma-Aldrich® | Custom |
|
nox5 ex4.4bp R: 5’-GCCTCAACATCAGCACCTAC-3’ |
Sigma-Aldrich® | Custom |
| M13 Reverse: 5’-GTAAAACGACGGCCAGTG-3’ |
Sigma-Aldrich® | Custom |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Enrique Amaya (enrique.amaya@manchester.ac.uk).
Materials availability
Plasmids generated in this study are available upon request from the lead contact. Zebrafish lines generated in this study will be deposited to the European Zebrafish Resource Centre.
Experimental model and subject details
The experimental model used for all in vivo experiments in this study was zebrafish (Danio rerio). All animal experiments were approved by the University of Manchester Animal Ethics Committee and were licenced in accordance with the Animals (Scientific Procedures) Act, 1986. Zebrafish husbandry was undertaken in a re-circulating system maintained at 28.5°C, with a 14hr photoperiod. These conditions are uniform for WT and all mutant strains. Embryos were obtained by marbling tanks, or by isolating pairs in breeding chambers. All experiments were carried out on random mixed sex cohorts. The age of the experimental animals used in this study ranged from 2 days post fertilisation (Figure S1) through to 22-month adults (Figures S3 and S4). Experiments focusing on comparing rate of caudal fin regeneration across different genotypes and/or HyPer imaging were carried out in adult animals between 6 and 8 months of age (Figures 3, 4 and 5, S2, S5-S8, S10 and S11). Data shown in Figure S9 was carried out in 3-month-old adults.
Method details
Genomic extraction
Fin clips from caudal fins of individual adults were added to a mixture of lysis Buffer (10mM Tris-HCL, pH 8.0, 1mM EDTA, 0.3% Tween-20, 0.3% NP40), and proteinase K (20-25mg/ml) (New England Biolabs® Inc.), 1ul/50ul lysis buffer. This was incubated in a thermal cycler programmed to 55°C (2hours), 95°C (10 minutes) and a 12°C hold.
sgRNA design and production of CRISPR mutants
Single guide RNAs (sgRNAs) were designed for targeting exon 1 of cyba, and exon 4 of nox5, as previously described (Moreno-mateos et al. 2015). Briefly, the Ensemble ID for each gene when entered online on CRISPRscan generated multiple gRNAs. Exon-targeting gRNAs were then chosen based on rank, location within the first 50% of the ORF, and distance from the initiation codon. A sgRNA template requires a 52nt oligo (sgRNA primer) 5’ TAATACGACTCACTATAGG (N = 18) GTTTTAGAGCTAGAA, containing the T7 promoter, the 20nt specific DNA-binding sequence [GG(N = 18)] and a constant 15nt tail annealing sequence. This was annealed to an invariant 80nt reverse oligo AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC 3′ tail primer, generating a 117bp PCR product. Oligos were obtained from Sigma-Aldrich®. The PCR cycler settings for this primer extension were 3 min at 95°C; 30 cycles of 30s at 95°C, 30s at 45°C and 30s at 72°C; and a final step at 72°C for 7 min. PCR products were purified using Qiaquick (Qiagen) columns. Approximately 120–150ng of DNA was used as a template for a T7 in vitro transcription reaction. In vitro transcribed sgRNAs were treated with DNase and precipitated using sodium acetate and ethanol. Purified sgRNA and Cas9-NLS protein (New England Biolabs® Inc.) were diluted to 300ng/ul. Equal volumes of Cas9-NLS protein, sgRNA and Phenol Red (Sigma-Aldrich®) were mixed to obtain the final injection mix. Injection drop size was adjusted to 1nl using a graticule scale. All embryos were injected at the one-cell stage. F1 heterozygous animals were identified via restriction digest, and indels were characterised using Sanger sequencing.
ENU mutant strains and transgenic strains
ENU mutants for cybasa11798 and duoxsa9892 were discovered during the Zebrafish Mutation Project (Kettleborough et al., 2013). These fish were sourced from the European Zebrafish Resource Centre (EZRC). The tshrwpr23e1 mutants (original name manetwpr23e1) were sourced from the laboratory of David Parichy (McMenamin et al. 2014). cybasa11798 and duoxsa9892 were identified via Sanger sequencing. The tshrwpr23e1 mutants were identified via restriction digest. The Tg(ubb:HyPer) reporter line (ubb promoter driving HyPer) was generated by Tol2 transgenesis as previously described.64 Mutants and transgenics generated in the lab were given allele designation via ZFIN.
Polymerase chain reaction
PCRs were undertaken using ExTaq DNA Polymerase (TaKaRa). Primers (Sigma Aldrich) used are listed below.
| cyba ex1.5bp F | Genomic PCR | 51.1°C |
| cyba ex1.5bp R | ||
| cyba sa11798 F | Genomic PCR | 54°C |
| cyba sa11798 R | ||
| duox sa9892 F | Genomic PCR | 55°C |
| duox sa9892 R | ||
| duox sa9892 seq | Sequencing | |
| manet F | Genomic PCR | 54°C |
| manet R | ||
| nox5 ex4.4bp F | Genomic PCR | 55°C |
| nox5 ex4.4bp R | ||
| M13 Reverse | Identifying indels |
Aspergillus fumigatus spore harvesting and microinjection
An Aspergillus fumigatus strain constitutively expressing the Turbo635 fluorescent protein in the A1163 background65 was used to infect the larvae. The strain was cultured on Aspergillus complete media (ACM) in a 20ml flask and incubated at 37°C for at least 2 days prior to spore harvesting. ACM (1L) was prepared using adenine (0.075g), glucose (10g), yeast extract (1g), bacteriological peptone (2g), casamino acids (1g), vitamin solution (10ml), salt solution (20ml), ammonium tartrate (10ml from 500mM stock), and 1.5% of agar. The pH was adjusted to 6.5 using 10M NaOH. Spores were harvested using a 0.5% NaCl / 0.002% Tween-20 solution (1X). 10ml of the 1X solution was added per flask and flasks were agitated to detach spores. This suspension was filtered through miracloth (Merck Millipore), followed by centrifugation at 4000RPM, for 5minutes. The resultant pellet was resuspended, followed again by centrifugation. Pellet was resuspended in 5ml of the 1X NaCl/Tween. 1/10 and 1/100 dilutions were made from this master solution using and were counted using a New Improved Neubauer haematocytometer (Marinfeld,Germany) on a Nikon Optiphot at a 40x magnification
Freshly harvested spores were used for each experiment. Prior to injection, larvae were anaesthetised in MS-222, and positioned laterally on injection plates made of 3% agarose in E3.66,67,68 A spore suspension containing 5 conidia/nl was directly injected into the yolk sac of 2dpf larvae, using a microinjection setup consisting of a Picospritzer II (General Valve Corporation) and a Leica MZ6 stereomicroscope. The injected volume was 1nl, unless specified otherwise. Viability of injected larvae was checked twice per day to monitor survival. Dead larvae were plated on Potato Dextrose Agar (PDA). Plates were incubated at 37°C and appearance of hyphal masses was used as confirmation for aspergillosis-led mortality.
Caudal fin amputation and regeneration
Fish were anaesthetised using 0.4 mg/mL (0.04%) MS-222 (Sigma Aldrich). Animals were imaged prior to amputation, at T0, 1, 2, 3, and 4weeks amputation (wpa), on a Leica M165 FC (Leica Microsystems). Using Photoshop CS5 (Adobe®), images were cropped at the level of the caudal peduncle, and fins were outlined using the brush tool. The “Record Measurements” command was used to obtain the total area of the outlined fin. Regeneration was calculated as a weekly increase, relative to the unamputated state, where the unamputated state was regarded as 100%. This is formulated as:
Animals that had less than 40% or more than 55% of the caudal fin amputated were excluded from further analysis, as we wished to eliminate any potential differences in the rates of regeneration to be caused by differences how much fin had been amputated as has been shown previously by others.39,40,41
H2O2 detection
For visualising H2O2 levels in the caudal fin, fish were imaged on a high-end widefield microscope (Decon Vision) using a 4X objective. MetaMorph® software was used to setup the mircroscope to capture two wavelengths - HyPer low (CFP) emission 482/25 and HyPer high (YFP) emission 544/24. Exposure was set 1000 microseconds and binning was 1X1. Excitation was achieved using BP 430/24 and BP 500/20 filters. Emission was detected with a BP 535/30 filter. Animals were imaged before and after amputation, over a series of time points. To do this, they were anaesthetised as routine, placed upon 24mm X 50mm glass coverslips in a thin film of anaesthetic and mounted. Using ImageJ images were analysed by subtracting background, smoothing, formatting to 32-bit, then dividing to calculate the HyPer ratio. The average H2O2 over the area captured in the image was then calculated and visualised using Prism 8.1 (GraphPad Software, Inc.).
Quantification and statistical analysis
MATLAB (MathWorks), SPSS (IBM Corporation) and GraphPad Prism 8.1 (GraphPad Software, Inc.) were used for all the statistical and computational tasks. All of the statistical details of experiments can be found in the figure legends. Null hypotheses were rejected when p value < 0.05. In all figure legends, n denotes the number of animals used for the experiment. The bar charts show the mean of the data with error bars representing the SD. The data was tested for normality, and normally distributed data was analysed using parametric tests (ANOVA and t-test). The measurements taken between different groups (WT and mutants) were independent. Equal variance of data was assumed as the residuals were found to be homoscedastic within groups. No outliers were found or excluded throughout the experiments.
Aspergillosis-related survival was statistically assessed using the Log Rank Test. For fin regeneration, comparisons were made using ANOVA (repeated measures and one-way) or t-test. Linear regressions were performed without interpolation, with a 95% significance level. The strength of the correlation was assessed by the correlation coefficient, adjusted R square. Where applicable, Pearson’s correlation coefficient and Spearman’s rank correlation coefficient was reported. For the unamputated fin, distribution was used for explaining the relationship between age and fin size. Oscillations were tested using power spectrum analysis based on Fourier transform as well as autocorrelation. MATLAB was used to perform power spectrum analysis and SSPS was used for autocorrelations.
Acknowledgments
We would like to thank the aquarium staff in the BSF unit for their care and support of the fish. We would also like to thank Kalin Narov, (https://kalinnarov.wixsite.com/embryosafari) for his contribution to Panel A in Figure 1, and Veronica Biga for her help with statistical methods. Special thanks go to Peter March, Roger Meadows, and Steven Marsden for their help with the microscopy. The Bioimaging Facility microscopes used in this study were purchased with grants from BBSRC, Wellcome, and the University of Manchester Strategic Fund. This work was supported by a PhD studentship from The Scar Free Foundation (K.C.), an MRC Research Project Grant (MR/L007525/1) (S.I., E.A.), and a grant from the British Heart Foundation Oxbridge Regenerative Medicine Centre (RM/17/2/33380) (K.C., E.A.).
Author contributions
Conceptualization: E.A., K.C., J.A.; Methodology: K.C., S.I., M.F., L.S., V.S., and R.B.; Validation: K.C.; Formal analysis: K.C.; Investigation: K.C., S.I., M.F., L.S., V.S., and R.B.; Writing - original draft: K.C.; Writing - review & editing: E.A.; Visualization: E.A.; Supervision: E.A. and J.A.; Project administration: E.A.; Funding acquisition: E.A.
Declaration of interests
The authors declare no conflicts of interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a member of the LGBTQIA + community.
Published: February 4, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106147.
Supplemental information
Data and code availability
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Wenger Y., Buzgariu W., Reiter S., Galliot B. Injury-induced immune responses in Hydra. Semin. Immunol. 2014;26:277–294. doi: 10.1016/j.smim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Gauron C., Meda F., Dupont E., Albadri S., Quenech’Du N., Ipendey E., Volovitch M., Del Bene F., Joliot A., Rampon C., Vriz S. Hydrogen peroxide (H2O2) controls axon pathfinding during zebrafish development. Dev. Biol. 2016;414:133–141. doi: 10.1016/j.ydbio.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Santabárbara-Ruiz P., López-Santillán M., Martínez-Rodríguez I., Binagui-Casas A., Pérez L., Milán M., Corominas M., Serras F. ROS-induced JNK and p38 signaling is required for unpaired cytokine activation during Drosophila regeneration. PLoS Genet. 2015;11:e1005595. doi: 10.1371/journal.pgen.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan S.J., Abidi S.N.F., Skinner A., Tian Y., Smith-Bolton R.K. The Drosophila Duox maturation factor is a key component of a positive feedback loop that sustains regeneration signaling. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Love N.R., Chen Y., Ishibashi S., Kritsiligkou P., Lea R., Koh Y., Gallop J.L., Dorey K., Amaya E. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat. Cell Biol. 2013;15:222–228. doi: 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai H., Zhang W., Qin X.-J., Zhang T., Wu H., Liu J.-Z., Hai C.-X. Hydrogen peroxide modulates the proliferation/quiescence switch in the liver during embryonic development and posthepatectomy regeneration. Antioxidants Redox Signal. 2015;22:921–937. doi: 10.1089/ars.2014.5960. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Q., Wang Y., Man L., Zhu Z., Bai X., Wei S., Liu Y., Liu M., Wang X., Gu X., Wang Y. Reactive oxygen species generated from skeletal muscles are required for gecko tail regeneration. Sci. Rep. 2016;6 doi: 10.1038/srep20752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al Haj Baddar N.W., Chithrala A., Voss S.R. Amputation-induced reactive oxygen species signaling is required for axolotl tail regeneration. Dev. Dynam. 2019;248:189–196. doi: 10.1002/dvdy.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pirotte N., Stevens A.-S., Fraguas S., Plusquin M., Van Roten A., Van Belleghem F., Paesen R., Ameloot M., Cebrià F., Artois T., Smeets K. Reactive oxygen species in planarian regeneration: an upstream necessity for correct patterning and brain formation. Oxid. Med. Cell. Longev. 2015;2015:392476. doi: 10.1155/2015/392476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romero M.M.G., McCathie G., Jankun P., Roehl H.H. Damage-induced reactive oxygen species enable zebrafish tail regeneration by repositioning of Hedgehog expressing cells. Nat. Commun. 2018;9:4010. doi: 10.1038/s41467-018-06460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sipka T., Peroceschi R., Hassan-Abdi R., Groß M., Ellett F., Begon-Pescia C., Gonzalez C., Lutfalla G., Nguyen-Chi M. Damage-induced calcium signaling and reactive oxygen species mediate macrophage activation in zebrafish. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.636585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hervera A., De Virgiliis F., Palmisano I., Zhou L., Tantardini E., Kong G., Hutson T., Danzi M.C., Perry R.B.-T., Santos C.X.C., et al. Reactive oxygen species regulate axonal regeneration through the release of exosomal NADPH oxidase 2 complexes into injured axons. Nat. Cell Biol. 2018;20:307–319. doi: 10.1038/s41556-018-0039-x. [DOI] [PubMed] [Google Scholar]
- 13.Gauron C., Rampon C., Bouzaffour M., Ipendey E., Teillon J., Volovitch M., Vriz S. Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed. Sci. Rep. 2013;3:2084. doi: 10.1038/srep02084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niethammer P., Grabher C., Look A.T., Mitchison T.J. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bedard K., Krause K.-H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 16.Reis J., Massari M., Marchese S., Ceccon M., Aalbers F.S., Corana F., Valente S., Mai A., Magnani F., Mattevi A. A closer look into NADPH oxidase inhibitors: Validation and insight into their mechanism of action. Redox Biol. 2020;32 doi: 10.1016/j.redox.2020.101466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ten Freyhaus H., Huntgeburth M., Wingler K., Schnitker J., Bäumer A.T., Vantler M., Bekhite M.M., Wartenberg M., Sauer H., Rosenkranz S. Novel Nox inhibitor VAS2870 attenuates PDGF-dependent smooth muscle cell chemotaxis, but not proliferation. Cardiovasc. Res. 2006;71:331–341. doi: 10.1016/j.cardiores.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Nachtrab G., Czerwinski M., Poss K.D. Sexually dimorphic fin regeneration in zebrafish controlled by androgen/GSK3 signaling. Curr. Biol. 2011;21:1912–1917. doi: 10.1016/j.cub.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azevedo A.S., Grotek B., Jacinto A., Weidinger G., Saúde L. The regenerative capacity of the zebrafish caudal fin is not affected by repeated amputations. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panday A., Sahoo M.K., Osorio D., Batra S. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell. Mol. Immunol. 2015;12:5–23. doi: 10.1038/cmi.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano Y., Longo-Guess C.M., Bergstrom D.E., Nauseef W.M., Jones S.M., Bánfi B. Mutation of the Cyba gene encoding p22phox causes vestibular and immune defects in mice. J. Clin. Invest. 2008;118:1176–1185. doi: 10.1172/jci33835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stasia M.J. CYBA encoding p22phox, the cytochrome b558 alpha polypeptide: gene structure, expression, role and physiopathology. Gene. 2016;586:27–35. doi: 10.1016/j.gene.2016.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jha J.C., Watson A.M.D., Mathew G., de Vos L.C., Jandeleit-Dahm K. The emerging role of NADPH oxidase NOX5 in vascular disease. Clin. Sci. 2017;131:981–990. doi: 10.1042/cs20160846. [DOI] [PubMed] [Google Scholar]
- 24.Kettleborough R.N.W., Busch-Nentwich E.M., Harvey S.A., Dooley C.M., de Bruijn E., van Eeden F., Sealy I., White R.J., Herd C., Nijman I.J., et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawahara T., Quinn M.T., Lambeth J.D. Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evol. Biol. 2007;7:109–121. doi: 10.1186/1471-2148-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chopra K., Ishibashi S., Amaya E. Zebrafish duox mutations provide a model for human congenital hypothyroidism. Biol. Open. 2019;8 doi: 10.1242/bio.037655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkelstein J.A., Marino M.C., Johnston R.B., Boyle J., Curnutte J., Gallin J.I., Malech H.L., Holland S.M., Ochs H., Quie P., et al. Chronic granulomatous disease: report on a national registry of 368 patients. Medicine. 2000;79:155–169. doi: 10.1097/00005792-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 28.Jones L.B.K.R., McGrogan P., Flood T.J., Gennery A.R., Morton L., Thrasher A., Goldblatt D., Parker L., Cant A.J. Special Article: chronic granulomatous disease in the United Kingdom and Ireland: a comprehensive national patient-based registry. Clin. Exp. Immunol. 2008;152:211–218. doi: 10.1111/j.1365-2249.2008.03644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martire B., Rondelli R., Soresina A., Pignata C., Broccoletti T., Finocchi A., Rossi P., Gattorno M., Rabusin M., Azzari C., et al. Clinical features, long-term follow-up and outcome of a large cohort of patients with Chronic Granulomatous Disease: an Italian multicenter study. Clin. Immunol. 2008;126:155–164. doi: 10.1016/j.clim.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Marciano B.E., Spalding C., Fitzgerald A., Mann D., Brown T., Osgood S., Yockey L., Darnell D.N., Barnhart L., Daub J., et al. Common severe infections in chronic granulomatous disease. Clin. Infect. Dis. 2015;60:1176–1183. doi: 10.1093/cid/ciu1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoen T.J., Rosowski E.E., Knox B.P., Bennin D., Keller N.P., Huttenlocher A. Neutrophil phagocyte oxidase activity controls invasive fungal growth and inflammation in zebrafish. J. Cell Sci. 2019;133 doi: 10.1242/jcs.236539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beauté J., Obenga G., Le Mignot L., Mahlaoui N., Bougnoux M.-E., Mouy R., Gougerot-Pocidalo M.-A., Barlogis V., Suarez F., Lanternier F., et al. Epidemiology and outcome of invasive fungal diseases in patients with chronic granulomatous disease. Pediatr. Infect. Dis. J. 2011;30:57–62. doi: 10.1097/inf.0b013e3181f13b23. [DOI] [PubMed] [Google Scholar]
- 33.Blumental S., Mouy R., Mahlaoui N., Bougnoux M.-E., Debré M., Beauté J., Lortholary O., Blanche S., Fischer A. Invasive mold infections in chronic granulomatous disease: a 25-year retrospective survey. Clin. Infect. Dis. 2011;53:e159–e169. doi: 10.1093/cid/cir731. [DOI] [PubMed] [Google Scholar]
- 34.Falcone E.L., Holland S.M. Invasive fungal infection in chronic granulomatous disease. Curr. Opin. Infect. Dis. 2012;25:658–669. doi: 10.1097/qco.0b013e328358b0a4. [DOI] [PubMed] [Google Scholar]
- 35.Collymore C., Tolwani A., Lieggi C., Rasmussen S. Efficacy and safety of 5 anesthetics in adult zebrafish (Danio rerio) J. Am. Assoc. Lab. Anim. Sci. 2014;53:198–203. [PMC free article] [PubMed] [Google Scholar]
- 36.Parichy D.M., Elizondo M.R., Mills M.G., Gordon T.N., Engeszer R.E. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev. Dynam. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chassot B., Pury D., Jaźwińska A. Zebrafish fin regeneration after cryoinjury-induced tissue damage. Biol. Open. 2016;5:819–828. doi: 10.1242/bio.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.König D., Jaźwińska A. Zebrafish fin regeneration involves transient serotonin synthesis. Wound Repair Regen. 2019;27:375–385. doi: 10.1111/wrr.12719. [DOI] [PubMed] [Google Scholar]
- 39.Uemoto T., Abe G., Tamura K. Regrowth of zebrafish caudal fin regeneration is determined by the amputated length. Sci. Rep. 2020;10:649. doi: 10.1038/s41598-020-57533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y., Grill S., Sanchez A., Murphy-Ryan M., Poss K.D. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- 41.Akimenko M.A., Johnson S.L., Westerfield M., Ekker M. Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development. 1995;121:347–357. doi: 10.1242/dev.121.2.347. [DOI] [PubMed] [Google Scholar]
- 42.McMenamin S.K., Bain E.J., McCann A.E., Patterson L.B., Eom D.S., Waller Z.P., Hamill J.C., Kuhlman J.A., Eisen J.S., Parichy D.M. Thyroid hormone-dependent adult pigment cell lineage and pattern in zebrafish. Science. 2014;345:1358–1361. doi: 10.1126/science.1256251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belousov V.V., Fradkov A.F., Lukyanov K.A., Staroverov D.B., Shakhbazov K.S., Terskikh A.V., Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 44.Taylor J.S., Braasch I., Frickey T., Meyer A., Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trubiroha A., Gillotay P., Giusti N., Gacquer D., Libert F., Lefort A., Haerlingen B., De Deken X., Opitz R., Costagliola S. A rapid CRISPR/Cas-based mutagenesis assay in zebrafish for identification of genes involved in thyroid morphogenesis and function. Sci. Rep. 2018;8:5647. doi: 10.1038/s41598-018-24036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grasberger H. Defects of thyroidal hydrogen peroxide generation in congenital hypothyroidism. Mol. Cell. Endocrinol. 2010;322:99–106. doi: 10.1016/j.mce.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 47.Itou J., Kawakami H., Burgoyne T., Kawakami Y. Life-long preservation of the regenerative capacity in the fin and heart in zebrafish. Biol. Open. 2012;1:739–746. doi: 10.1242/bio.20121057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wendler S., Hartmann N., Hoppe B., Englert C. Age-dependent decline in fin regenerative capacity in the short-lived fish Nothobranchius furzeri. Aging Cell. 2015;14:857–866. doi: 10.1111/acel.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Razzell W., Evans I.R., Martin P., Wood W. Calcium flashes orchestrate the wound inflammatory response through DUOX activation and hydrogen peroxide release. Curr. Biol. 2013;23:424–429. doi: 10.1016/j.cub.2013.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soto X., Li J., Lea R., Dubaissi E., Papalopulu N., Amaya E. Inositol kinase and its product accelerate wound healing by modulating calcium levels, Rho GTPases, and F-actin assembly. Proc. Natl. Acad. Sci. USA. 2013;110:11029–11034. doi: 10.1073/pnas.1217308110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panda S. Circadian physiology of metabolism. Science. 2016;354:1008–1015. doi: 10.1126/science.aah4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Honkoop H., de Bakker D.E., Shoffner A., et al. Aharonov A., Kruse F., Shakked A., Nguyen P.D., de Heus C., Garric L., et al. Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. Elife. 2019;8 doi: 10.7554/elife.50163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love N.R., Ziegler M., Chen Y., Amaya E. Carbohydrate metabolism during vertebrate appendage regeneration: what is its role? How is it regulated?: a postulation that regenerating vertebrate appendages facilitate glycolytic and pentose phosphate pathways to fuel macromolecule biosynthesis. Bioessays. 2014;36:27–33. doi: 10.1002/bies.201300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinclair J.W., Hoying D.R., Bresciani E., Nogare D.D., Needle C.D., Berger A., Wu W., Bishop K., Elkahloun A.G., Chitnis A., et al. The Warburg effect is necessary to promote glycosylation in the blastema during zebrafish tail regeneration. NPJ Regen. Med. 2021;6:55. doi: 10.1038/s41536-021-00163-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brandão A.S., Borbinha J., Pereira T., Brito P.H., Lourenço R., Bensimon-Brito A., Jacinto A. Zebrafish caudal fin amputation induces a metabolic switch necessary for cell identity transitions and cell cycle re-entry to support blastema formation and bone regeneration. bioRxiv. 2022 doi: 10.1101/2022.01.26.477895. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott C.A., Carney T.J., Amaya E. Aerobic glycolysis is important for zebrafish larval wound closure and tail regeneration. Wound Repair Regen. 2022;30:665–680. doi: 10.1111/wrr.13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoyle N.P., Seinkmane E., Putker M., Feeney K.A., Krogager T.P., Chesham J.E., Bray L.K., Thomas J.M., Dunn K., Blaikley J., O'Neill J.S. Circadian actin dynamics drive rhythmic fibroblast mobilization during wound healing. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue Y., Liu P., Wang H., Xiao C., Lin C., Liu J., Dong D., Fu T., Yang Y., Wang Z., et al. Modulation of circadian rhythms affects corneal epithelium renewal and repair in mice. Invest. Ophthalmol. Vis. Sci. 2017;58:1865–1874. doi: 10.1167/iovs.16-21154. [DOI] [PubMed] [Google Scholar]
- 59.Sandvig K.U., Haaskjold E., Refsum S.B. Time dependency in the regenerative response to injury of the rat corneal epithelium. Chronobiol. Int. 1994;11:173–179. doi: 10.3109/07420529409057237. [DOI] [PubMed] [Google Scholar]
- 60.Brown S.A. Circadian clock-mediated control of stem cell division and differentiation: beyond night and day. Development. 2014;141:3105–3111. doi: 10.1242/dev.104851. [DOI] [PubMed] [Google Scholar]
- 61.Kowalska E., Ripperger J.A., Hoegger D.C., Bruegger P., Buch T., Birchler T., Mueller A., Albrecht U., Contaldo C., Brown S.A. NONO couples the circadian clock to the cell cycle. Proc. Natl. Acad. Sci. USA. 2013;110:1592–1599. doi: 10.1073/pnas.1213317110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.El-Brolosy M.A., Kontarakis Z., Rossi A., Kuenne C., Günther S., Fukuda N., Kikhi K., Boezio G.L.M., Takacs C.M., Lai S.-L., et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019;568:193–197. doi: 10.1038/s41586-019-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poburko D., Santo-Domingo J., Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J. Biol. Chem. 2011;286:11672–11684. doi: 10.1074/jbc.m110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love N.R., Thuret R., Chen Y., Ishibashi S., Sabherwal N., Paredes R., Alves-Silva J., Dorey K., Noble A.M., Guille M.J., et al. pTransgenesis: a cross-species, modular transgenesis resource. Development. 2011;138:5451–5458. doi: 10.1242/dev.066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Macdonald D., Thomson D.D., Johns A., Contreras Valenzuela A., Gilsenan J.M., Lord K.M., Bowyer P., Denning D.W., Read N.D., Bromley M.J. Inducible cell fusion permits use of competitive fitness profiling in the human pathogenic fungus Aspergillus fumigatus. Antimicrob. Agents Chemother. 2019;63:e01615–e01618. doi: 10.1128/aac.01615-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brothers K.M., Newman Z.R., Wheeler R.T. Live imaging of disseminated candidiasis in zebrafish reveals role of phagocyte oxidase in limiting filamentous growth. Eukaryot. Cell. 2011;10:932–944. doi: 10.1128/ec.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Knox B.P., Deng Q., Rood M., Eickhoff J.C., Keller N.P., Huttenlocher A. Distinct innate immune phagocyte responses to Aspergillus fumigatus conidia and hyphae in zebrafish larvae. Eukaryot. Cell. 2014;13:1266–1277. doi: 10.1128/ec.00080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rosowski E.E., Raffa N., Knox B.P., Golenberg N., Keller N.P., Huttenlocher A. Macrophages inhibit Aspergillus fumigatus germination and neutrophil-mediated fungal killing. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.