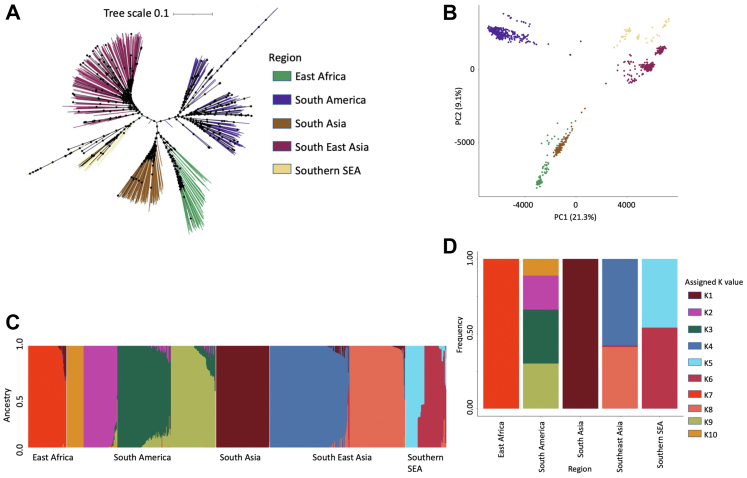
Fig. 1.
Population structure of 885 P. vivax isolates from 26 countries. A) Maximum likelihood phylogenetic tree generated using IQTREE from the pairwise SNP matrix of the complete global dataset of 885 samples and 454,681 SNPs. IQTREE was run using ModelFinder, tree search, ultrafast bootstrap and SH-aLRT test. Bootstrap scores between 50% and 100% are annotated on the tree branches with a black circle. Branches are colouring according to regional grouping (East Africa, n = 84, green branches; South America, n = 315, purple branches; South Asia, n = 114, brown branches; Southeast Asia (SEA), n = 286, dark pink branches; Southern SEA, n = 86, yellow branches). The phylogenetic tree file was visualised in iTOL with midpoint rooting. B) Principal component (PC) analysis displaying principal components 1 and 2 of the distance matrix generated using the SNP matrix. Each point represents and individual sample, coloured according with the region assigned in (A). PC 1 summarises 21.3% of the total variation whilst principal component 2 summarises 9.1% of the total variation. C) ADMIXTURE analysis of the global dataset predicted a total of 10 ancestral populations spread across each region: East Africa, n = 1; South America, n = 4; South Asia, n = 1; SEA, n = 3; SSEA, n = 2. D) Bar plot summarising the number and proportion of each ancestral population within each region.