Abstract
The combination of traditional Chinese medicine (TCM) and Western medicine is a promising method for treating rheumatoid arthritis (RA). Combining the two fully exploits the advantages of Western and TCM to treat RA and has the potential to greatly improve the therapeutic effect on RA. In this study, we developed a combination drug training set by using 16 characteristic variables based on the characteristics of small molecules of TCM ingredients and Food and Drug Administration-certified combination drug data downloaded from the DrugCombDB database. Furthermore, we compared the prediction and classification abilities of five models: the k-nearest neighbors, naive Bayes, support vector machine, random forest, and AdaBoost algorithms. The random forest model was selected as the classification and prediction model for Western and TCM and Western combination drugs. We collected data for 41 small molecules of TCM ingredients from the Traditional Chinese Medicine Systems Pharmacology database and 10 small molecule drugs commonly used in anti-RA treatment from the DrugBank database. Combinations of Western and TCM for anti-RA treatment were screened. Finally, the CellTiter-Glo method was used to determine the synergy of these combinations, and the 15 most predicted drug combinations were carried out experimental verification. Myricetin, rhein, nobiletin, and fisetin had high synergy with celecoxib, and rhein had high synergy with hydroxychloroquine. The preliminary findings of this study can be further applied for practical clinical anti-RA combined treatment strategies and serve as a reference for clinical treatment of RA with integrated Western and TCM.
1. Introduction
Modern medical research has demonstrated that many diseases often present complex pathogenic mechanisms, and consequently, the past methods for treating diseases with drugs are no longer adequate. In recent years, drug combination has been reported to offer clear advantages in treating tumors, AIDS, cardiovascular diseases, osteoarthritis, and other complex diseases; it has been widely implemented in clinical practice [1–4], with treatment methods based on combined drugs demonstrating advantageous synergistic effects, or a “1 + 1 ≥ 2” effect. The therapeutic effects of two or more drug combinations are greater than those achieved when each drug is used alone. Therefore, developing and evaluating potentially synergetic combination drugs, whether for development of new drugs or clinical treatment optimization, are essential. However, most existing combination drugs have been developed on the basis of limited clinical experience or experimental strategies. Verified combination drugs with synergistic therapeutic effects currently comprise a small portion of listed drugs. Therefore, making full use of knowledge of existing drug combinations, discovering new drug combinations based on well-accepted and understood drugs, and establishing a model for evaluating the synergistic effects of drug combinations are key concerns that must be addressed.
Rheumatoid arthritis (RA) is a chronic, systemic, inflammatory disease of the synovial and other joints. RA is a chronic progressive autoimmune disease [5], and its underlying pathogenesis is currently unclear. However, studies have reported that the incidence of RA increases with age due to infection and inflammatory mediators, and approximately 0.3%–1.0% of the global population is affected by the disease every year [6, 7]. At present, the drugs used in clinical treatment of RA are mainly nonsteroidal anti-inflammatory drugs (NSAIDs), disease modifying antirheumatic drugs (DMARDs), and biological macromolecular therapy. Because RA is complex, several studies have explored different combination therapy strategies for its treatment [8–10]. However, some therapeutic drugs, such as methotrexate, may produce unfavorable side effects [11, 12]. Therefore, if sole use of anti-RA combination therapy has the potential to produce unfavorable side effects, long-term use in patients with RA may do greater harm. Traditional Chinese medicine (TCM) has long been used to treat RA, and several studies have found that combined anti-RA TCM and a compound prescription have not only anti-inflammatory, analgesic, immune regulation, and multilevel and multilink therapeutic effects but also the advantages of high safety, few adverse reactions, and low cost. Such a combination has gained attention in studies of RA treatment and has increasingly attracted international attention [13, 14].
Therefore, we propose that the combination of Western medicine and TCM may be a promising method for improving the efficacy of anti-RA treatments and reducing side effects. Through their combination, the advantages of both Western medicine and TCM can be fully exploited to improve the therapeutic effects of RA treatment. However, because numerous active ingredients in TCM and hundreds of listed Western medicines have been reported to be effective in anti-RA treatments, the potential drug combinations are in the thousands or even tens of thousands. This multitude of combinations cannot feasibly be evaluated or verified through clinical practice and trials; therefore, the most practical method for identifying potential combinations would be to use drug data for existing combination drugs to model, evaluate, and screen potential anti-RA combinations of Western medicine and TCM and then experimentally validate these models. Therefore, in this study, we developed a classification and prediction model for combining Western and TCM drugs based on characteristics obtained through small molecule research on TCM and existing data on combined drugs. This model was developed using machine learning modeling followed by manual screening of anti-RA combination drugs. Through experimental verification, we identified effective combinations of Western and TCM drugs for treating RA; these findings can serve as a reference for clinical treatment of RA by using such combined treatment. The flow chart illustrating the process of drug discovery and verification for anti-RA integrative medicine through machine learning modeling is displayed in Figure 1.
Figure 1.
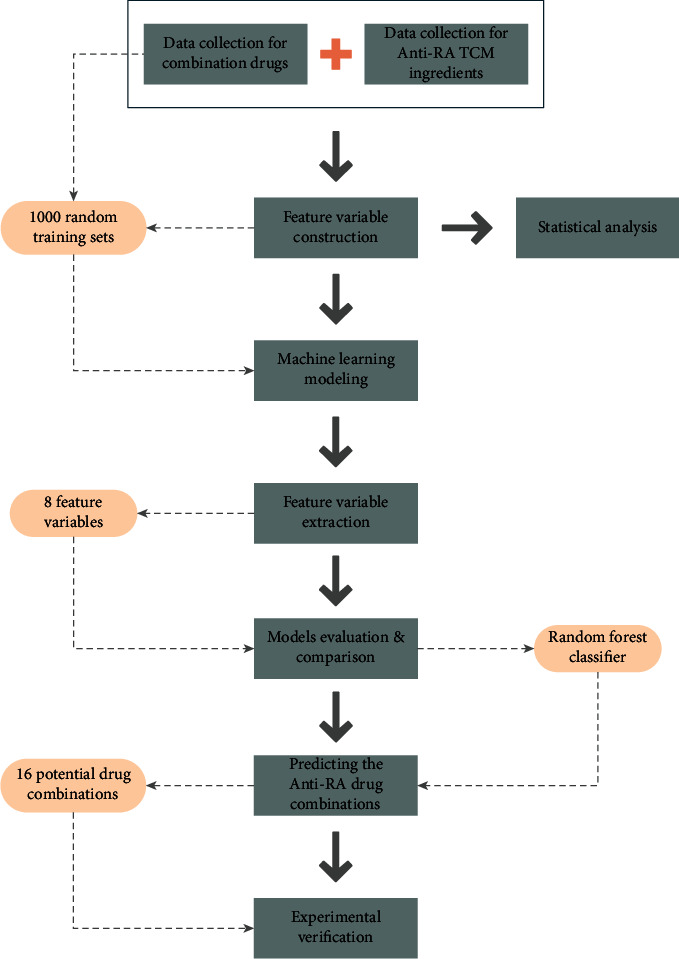
Flow chart of drug discovery and verification for anti-RA integrative medicine through machine learning modeling.
2. Materials and Methods
2.1. Data Collection and Collation
The DrugCombDB database (http://drugcombdb.denglab.org/) is a comprehensive combinatorial drug database [15] containing integrated drug combinations from various data sources. To ensure the reliability of the research data, we first downloaded the Food and Drug Administration- (FDA-) provided known drug combination dataset from the database (up to May 31, 2019). The molecular formula, Chemical Abstracts Service (CAS) number, canonical simplified molecular-input line-entry system (SMILES), and other chemical informatics data corresponding to each combination of drugs and their action targets were collected from the DrugBank database (https://go.drugbank.com/).
Furthermore, we searched “Rheumatoid Arthritis” on the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database (https://tcmspw.com/) to retrieve target Chinese medicine ingredient data reported to be relevant to treating RA. We obtained the main small molecules chemical information from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). In addition, several common anti-RA drugs were selected from the DrugBank database along with those discussed in the literature and those recommended by clinicians. All small molecule data for the TCM ingredients (collected from the TCMSP) and for known anti-RA Western drugs were combined in pairs to form the final prediction sample set.
2.2. Construction of Combined Drug Characteristic Variables
2.2.1. Drug Combination Features Based on Molecular Fingerprint Similarity
Canonical SMILES of all drugs were collected from the DrugBank and PubChem databases, and the 1024-dimensional molecular fingerprints for each of the small molecule drugs were calculated using DRAGON software (version 7.0). The fingerprint similarity (SFP) between two small molecule drugs was then calculated using Tanimoto coefficient-based similarity:
| (1) |
where A is the 1024-dimensional molecular fingerprint feature vector corresponding to the small molecule drug dA, ‖A‖ is the vector length, B is the 1024-dimensional molecular fingerprint feature vector corresponding to the small molecule drug dB, ‖B‖ is the vector length, and A∙B is the vector inner product.
2.2.2. Drug Combination Characteristics Based on Small Molecule ADME Similarity
To predict the adsorption, distribution, metabolism, and excretion (ADME) characteristics of each drug, Canonical SMILES for all small molecule drugs were imported into the SwissADME online system (http://www.swissadme.ch/) [16]. We selected 12 indicators as characteristic indexes for our ADME similarity calculations: molecular weight, heavy atoms, aromatic heavy atoms, fraction Csp3, rotatable bonds, H-bond acceptors, H-bond donors, molecular refractivity, topological polar surface area, Silicos-IT Log P, and Silicos-IT LogSw. All ADME indexes were first normalized, and the Euclidean distance was then used to calculate the similarity value SADME of ADME:
| (2) |
where xiA is the ADME index corresponding to the drug small molecule dA and xiB is the ADME index corresponding to the drug small molecule dB (n = 12).
2.2.3. Drug Combination Characteristics Based on Sequence Similarity of Small Molecular Targets
The action targets of all drugs were obtained from DrugBank and the TCMSP, and the protr package (version 1.6.2) based on R language was used to obtain the sequence of action protein targets of all small molecule drugs. The sequence similarity between the two target proteins, SSeq〈dA, dB〉, was calculated using the Smith–Waterman algorithm. The maximum, minimum, median, and mean values of the sequence similarity SSeq〈dA, dB〉 between all target proteins of the dA and dB were included in the characteristic variables as SSeqmax, SSeqmin, SSeqmed, and SSeqmean, respectively.
2.2.4. Drug Combination Characteristics Based on GO Functional Similarity of Small Molecular Targets
To calculate the gene ontology (GO) functional similarity SGO〈dA, dB〉 of the two drugs dA and dB, we used the GOSemSim package (version 2.14.2). Similarly, the minimum, maximum, median, and mean values of the GO functional similarity SGO〈dA, dB〉 of the two drugs dA and dB were included in the characteristic variables as SGOmin, SGOmax, SGOmed, and SGOmean, respectively.
2.2.5. Drug Combination Characteristics Based on Pathway Similarity of Small Molecule Targets
We used the clusterProfiler package (version 3.14.3) to perform a Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway enrichment analysis on all drug small molecule targets. We then screened the enrichment pathways of all targets based on p value < 0.05 and q value < 0.05. A vector, P = (p1, p2, ⋯, pn), consisting of all pathways was constructed according to the enriched pathways, {pk|k = 1, 2, ⋯, n} (where n is the number of pathways). If a target was enriched in a pathway pk in the target set TA of the drug small molecule dA, then the value was set as pkA = 1 at the corresponding position of the drug small molecule A pathway vector PA, and if not, it was set as 0:
| (3) |
The target pathway vector PB of the drug small molecule dB can be similarly obtained using a string similarity formula to calculate the pathway similarity value (SPathway) of two drugs:
| (4) |
where ‖PA‖ and ‖PB‖ are path vector lengths and PA · PB is the vector inner product.
2.2.6. Drug Combination Characteristics Based on Distance of Small Molecule Targets in Human PPI Network
The distance and network proximity of small molecule targets of different combinations of drugs on the human protein–protein interaction (PPI) network were calculated to measure the synergy characteristics of combination drugs within the PPI network:
| (5) |
where d (a, b) is the shortest path distance between two protein targets, a and b, within the PPI network (calculated using the igraph package [version 1.2.6]), 〈dAB〉 = 1/‖A‖ × ‖B‖∑a∈A, b∈Bd(a, b), ‖A‖ is the number of targets for the drug small molecule dA, and ‖B‖ is the number of targets for the drug small molecule dB. The human PPI background network used in the calculation was derived from the literature [17]; the PPI network contained a total of 16677 proteins and 243603 interactions.
The Tanimoto coefficient-based similarity, Euclidean distance, and cosine similarity calculations used in this paper were calculated using the philentropy package (version 0.4.0) based on R language (version 4.0.2).
2.3. Construction and Selection of Combination Drug Prediction Model
To comprehensively determine and select an appropriate machine learning model for modeling and prediction in this study, we selected different machine learning classifiers for use with the training set and the verification set and compared their classification accuracies. In this study, we selected and compared five classical machine learning models: the k-nearest neighbors (k-NN) [18], naive Bayes (NB) [19], support vector machine (SVM) [20], random forest (RF) [21], and AdaBoost classifiers [22]. The caret package (version 6.0-86), the klaR package (version 0.6-15), the svmRadial model in the kernlab package (version 0.9-29), the randomForest package (version 4.6-14), and the ada package (version 2.0-5) were used for the k-NN, naive Bayes, support vector machine, random forest, and AdaBoost classifiers, respectively.
Furthermore, we used established sample training sets to compare the classification prediction effects of the five machine learning models. First, the sample set was divided into prediction training sets and internal validation sets at a 7 : 3 ratio. Then, the prediction training set was input into the five machine learning models, and the machine learning models were trained using three sets of 10-fold cross-validation. The trained machine learning models were evaluated using the internal validation sets, and the model with the best classification prediction effect was selected as the final drug prediction model for the Western and TCM drug combinations.
2.4. Evaluation of Machine Learning Models
In this study, we defined precision rate as the proportion of positive samples correctly predicted by the model out of all positive predictions, recall rate as the proportion of positive samples correctly predicted by the model out of all predictions, accuracy as the proportion of positive and negative samples to all samples correctly predicted by the model, and F1-score as the overall score of the model when precision rate and recall rate were given equal weight (calculated using Formulas (6)–(9), respectively). A higher score indicated a better classification effect.
| (6) |
| (7) |
| (8) |
| (9) |
where true positive (TP) indicates a sample is positive and was predicted to be positive, false negative (FN) indicates a sample is positive but was predicted to be negative, false positive (FP) indicates a sample is negative but was predicted to be positive, and true negative (TN) indicates a sample is negative and was predicted to be negative.
In the plot of the receiver operating characteristic (ROC) curve, the horizontal axis represents the false positive rate (FPR) and the vertical axis represents the true positive rate (TPR). The area under the ROC curve is the area surrounded by the ROC and its lower coordinate axis and is represented as AUROC in this paper. In addition, because the precision and recall rates of the model are not fixed, when the classification threshold of the model is adjusted, the precision and recall rates continue to change. The precision–recall curve represents the relationship between the precision rate of the ordinate and the recall rate of the abscissa with the change of the classification threshold p value; the area beneath this curve is represented as the area under the precision–recall curve (AUPRC).
2.5. In Vitro Cell Experiment Verification
2.5.1. Cell Culture and Reagents
RAW264.7 cells were purchased from the American Type Culture Collection. The culture reagents included Dulbecco's modified Eagle medium (DMEM; Gibco, C11995500CP), Roswell Park Memorial Institute (RPMI) 1640 (Gibco, C11875500BT), fetal bovine serum (Bio IND, 04-002-1A), antimycotic (Lifetechnologies, 15240-112), phosphate-buffered saline (PBS), pH 7.4 (Gibco, 10010-023), trypsin-EDTA (0.05%; Lifetechnologies, 25300-054), bovine serum albumin (Lifetechnologies, 15561012), and CellTiter-Glo (CTG; PROMEGA, G7572). Consumables and instruments included a cell culture plate (Corning), cell culture flask (Corning), microplate tester (BioTek, HM-1), and conventional instruments, such as a CO2 incubator (Thermo 3111) and biosafety cabinet (Heal Force, HFSAFe-1200LC).
2.5.2. Experimental Procedure and Methods
First, the different compounds were diluted with DMEM medium to the desired concentrations of the experimental design, namely, 0, 0.032, 0.16, 0.8, 4, 20, and 100 μM. The cells were then counted and inoculated into 96-well culture plates at 2000 cells/100 μL/well; 90 μL of culture medium (including serum) was added to each well for overnight culture, and 10 μL of drug solution was added. After a 48 h culture, CTG was used to detect the proliferation of RAW264.7 cells; CTG (100 μL) was added to each well, the content of which was incubated at room temperature in a dark environment for 10 min. The chemiluminescence value of each well was measured at 500 ms using enzyme calibration.
2.5.3. Combination Index Measurement
In this study, according to the principle of the median effect—specifically by employing the Chou–Talalay model [1]—the synergistic effect between each set of drugs was analyzed, and the effects of integrated Western and TCM drugs on RA cell lines were evaluated using the combination index (CI). The CI indicates the degree of drug synergy, addition, or antagonism for any given drug combination:
| (10) |
where when CI = 1, the two drugs have an additive effect, when CI < 1, the two drugs have a synergistic effect, and when CI > 1, the two drugs are antagonistic. DA and DB represent the concentrations of the two drugs, dA and dB, when administered in combination, and (DA)x and (DB)x represent the concentrations of the two drugs, dA and dB, when they have been administered alone and the combined drug inhibition rate has been reached.
| (11) |
In Formula (11), fa and fu are the inhibition and survival rates, respectively, when the two drugs are combined, fa = 1 − fu, Dm is the median dose, and m is the curve coefficient (calculated according to half-maximal inhibitory concentration (IC50) theory).
In this study, CompuSyn software (http://www.combosyn.com/) was used to calculate the CI values of the drug combinations to evaluate whether each screened combination of Western and TCM drugs had synergistic therapeutic effects.
2.6. Statistical Analysis
Data were expressed as the mean ± standard deviation, and a t-test and Mann–Whitney nonparametric tests were performed using IBM SPSS Statistics (version 26.0).
3. Results
3.1. Collection and Collation of Combined Drug Data
We obtained 946 pairs (comprising 816 drugs) of combinatorial drug data reported by the FDA from the DrugCombDB database. We further downloaded all existing small molecule drug data from the DrugBank database. After name matching and sorting, some small molecule drug data without key information, such as targets, were removed. In total, we included 488 drugs and their corresponding 546 pairs of combinations. The data obtained for each small molecule drug mainly included DrugBank ID, canonical SMILES, and known targets (Supplementary 1; pretreated FDA-certified combination drug dataset).
3.2. Construction of Combination Drug Training Set
First, using the method described in Section 2.2, we obtained the molecular fingerprint similarity value (SFP), the ADME index similarity value (SADME), and the minimum, maximum, median, and mean of the similarity (SSeqmin, SSeqmax, SSeqmed, and SSeqmean, respectively) of the drug target sequence between the two small molecule drugs. The minimum, maximum, median, and mean values of the GO functional similarity of the drug targets were SGOmin, SGOmax, SGOmed, and SGOmean, respectively. The KEGG pathway similarity value (SPathway); the minimum, maximum, median, and mean of the shortest path of different drug targets on the human PPI network (SPPImin, SPPImax, SPPImed, and SPPImean, respectively); and the proximity (Snp) of the different drug targets were also obtained. Thus, we obtained a total of 16 characteristic variables to predict combinations of Western and TCM drugs.
In this study, we evaluated 16 characteristic variables between two drugs for 546 pairs of combined drugs and removed those for which characteristic variables could not be calculated or were missing. We obtained a total of 404 positive samples of combined drugs (comprising 488 drugs) reported by the FDA. Due to a lack of effective noncombination drug data, we combined the 488 drugs in pairs, removed the existing combination drug samples and those missing data, and obtained a total of 45603 noncombination drugs as negative samples (Supplementary 2; positive and negative sample training dataset for all drug combinations).
Finally, to construct the training set for the TCM-combined drug samples, because the number of noncombined drugs should be much larger than that of combined drugs and to avoid large deviation in positive and negative samples, we set the ratio of positive to negative samples as 1 : 5; the number of positive samples was 404, and the number of negative samples randomly extracted from the 45603 noncombined drug samples was 404 × 5 = 2020. Through this, a training sample set was formed containing 2424 samples, and the training set was repeated 1000 times to randomly generate 1000 prediction training sets of combined drugs, as displayed in Figure 2.
Figure 2.
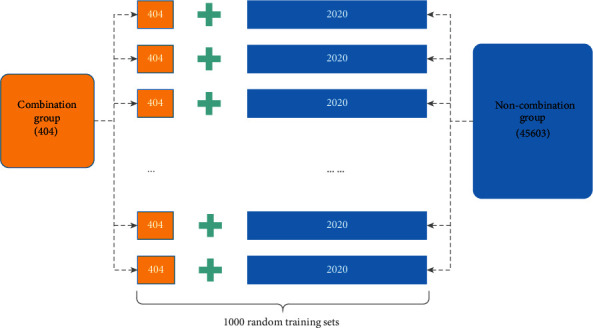
Training set of prediction samples comprised 1000 training sets of prediction samples randomly selected from 2020 samples of noncombination drugs and 404 samples of combination drugs.
3.3. Analysis and Screening of Characteristic Variables in Combination Drug Training Set
A t-test and a Mann–Whitney nonparametric test were used to determine whether significant differences existed between the 16 characteristic variables in the combined drug group and those in the noncombined drug group (p < 0.05). The results revealed that SFP, SADME, SSeqmin, SGOmax, SGOmed, SGOmean, and SPPImax did not significantly differ between the combined drug and noncombined drug groups (p > 0.05). Significant differences were found in the SSeqmax, SSeqmed, SSeqmean, SPathway, SPPImin, SPPImed, SPPImean, and Snp between the two statistical tests (p < 0.05). Statistical differences were found in SGOmin in the nonparametric tests (p < 0.05). All statistical tests were performed in IBM SPSS Statistics (version 26.0), and their data are presented in Table 1. The violin diagrams of the distribution of characteristic variables in different groups were completed using ggplot2 (version 3.3.5) and are displayed in Figure 3.
Table 1.
Results of t-test and Mann–Whitney nonparametric test for 16 characteristic variables between different groups.
| Feature | Noncombination group | Combination group | t-test | Mann–Whitney test |
|---|---|---|---|---|
| S FP | 0.783 ± 0.087 | 0.783 ± 0.091 | 0.914 | 0.664 |
| S ADME | 3.347 ± 2.994 | 3.160 ± 2.697 | 0.167 | 0.199 |
| S Seqmin | 0.014 ± 0.045 | 0.020 ± 0.070 | 0.109 | 0.400 |
| S Seqmax | 0.066 ± 0.189 | 0.173 ± 0.342 | ∗∗ | ∗∗ |
| S Seqmed | 0.021 ± 0.057 | 0.030 ± 0.084 | ∗ | ∗∗ |
| S Seqmean | 0.026 ± 0.062 | 0.049 ± 0.105 | ∗∗ | ∗∗ |
| S GOmin | 0.236 ± 0.175 | 0.221 ± 0.193 | 0.114 | ∗∗ |
| S GOmax | 0.523 ± 0.239 | 0.550 ± 0.288 | 0.061 | 0.092 |
| S GOmed | 0.374 ± 0.172 | 0.369 ± 0.205 | 0.649 | 0.532 |
| S GOmean | 0.376 ± 0.166 | 0.374 ± 0.194 | 0.825 | 0.644 |
| S Pathway | 0.007 ± 0.029 | 0.014 ± 0.039 | ∗∗ | ∗∗ |
| S PPImin | 0.867 ± 0.092 | 0.821 ± 0.142 | ∗∗ | ∗∗ |
| S PPImax | 0.931 ± 0.116 | 0.934 ± 0.094 | 0.500 | 0.889 |
| S PPImed | 0.917 ± 0.043 | 0.910 ± 0.050 | ∗∗ | ∗∗ |
| S PPImean | 0.100 ± 0.064 | 0.121 ± 0.091 | ∗∗ | ∗∗ |
| S np | −0.487 ± 0.215 | −0.436 ± 0.216 | ∗∗ | ∗∗ |
∗p < 0.05 and∗∗p < 0.01.
Figure 3.
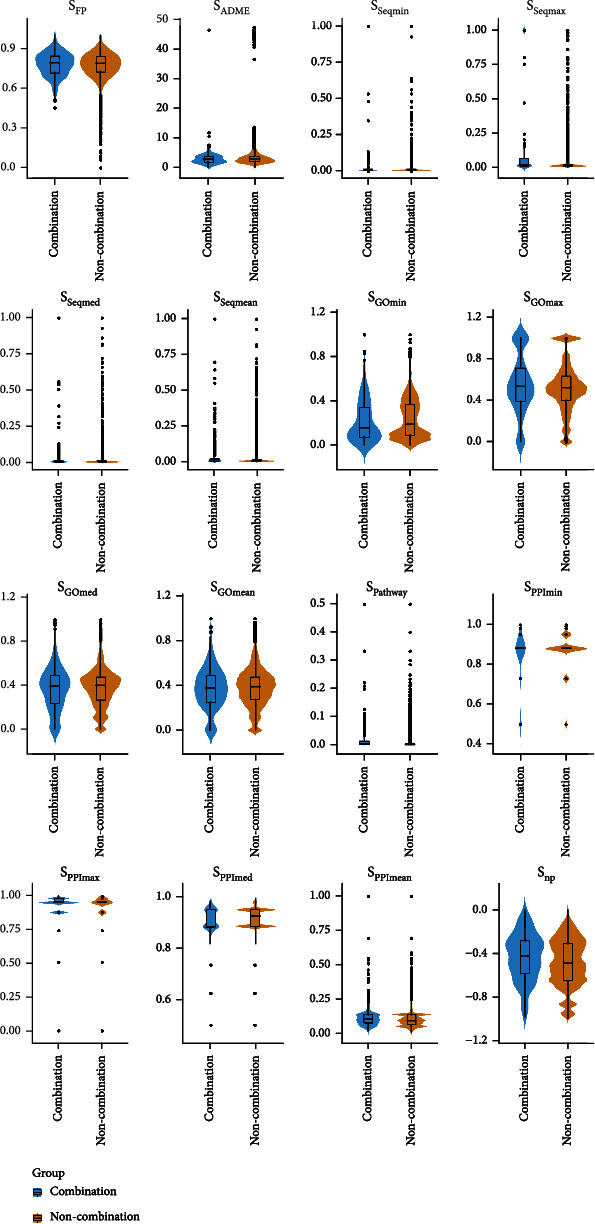
Violin diagram of value distribution of 16 characteristic variables in training sample set.
An rfe function using the caret package was used to screen the 16 characteristic variables. A random forest model was used as the selection function, which was functions = rfFuncs, and the method was cross-validation, which was method =“cv.” The others parameters were set to default. The training set was repeated 1000 times to extract feature variables from 1000 randomly generated training set samples, and each extracted feature variable set was recorded. The frequency of each feature variable in the 1000 feature selections was counted; the results are displayed in Figure 4.
Figure 4.
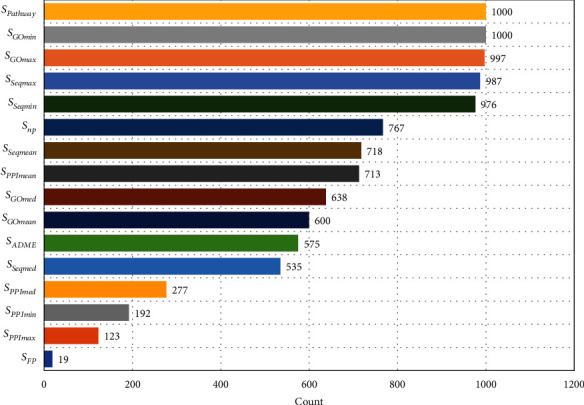
Occurrence of 16 feature variables from 1000 feature extractions.
As illustrated in Figure 4, eight characteristic variables, namely, SPathway, SGOmin, SGOmax, SSeqmin, SSeqmax, SSeqmean, SPPImean, and Snp, had appearance frequencies of over 70% after 1000 characteristic screenings. SPathway and SGOmin were identified as key characteristic variables for predicting combined drugs. The other eight indicators had appearance frequencies below 70%; for example, SFP had a rate of less than 10%. Thus, we removed the group of indicators with frequencies lower than 70% and included the top eight indicators as feature variables for subsequent machine learning.
3.4. Comparison and Selection Results of Machine Learning Models
The five included machine learning models were subjected to three replicates of 10-fold cross-validation under 1000 random samples to evaluate the predictive classification performance of different models. The results are displayed in Table 2, and the ROC and PR curves of the five models are presented in Figure 5.
Table 2.
Prediction evaluation results of five machine learning classification models under 1000 random sample training sets.
| Model | Accuracy | Precision | Recall | F1 score | AUROC | AUPR |
|---|---|---|---|---|---|---|
| k-NN | 0.827 ± 0.012 | 0.848 ± 0.012 | 0.965 ± 0.013 | 0.903 ± 0.008 | 0.651 ± 0.024 | 0.309 ± 0.033 |
| NB | 0.813 ± 0.014 | 0.845 ± 0.012 | 0.949 ± 0.013 | 0.894 ± 0.009 | 0.617 ± 0.026 | 0.269 ± 0.031 |
| SVM | 0.834 ± 0.011 | 0.838 ± 0.012 | 0.992 ± 0.006 | 0.909 ± 0.007 | 0.657 ± 0.027 | 0.330 ± 0.038 |
| RF | 0.849 ± 0.012 | 0.861 ± 0.012 | 0.977 ± 0.008 | 0.915 ± 0.007 | 0.739 ± 0.024 | 0.460 ± 0.039 |
| AdaBoost | 0.833 ± 0.011 | 0.834 ± 0.012 | 0.998 ± 0.004 | 0.909 ± 0.007 | 0.667 ± 0.027 | 0.317 ± 0.038 |
Figure 5.
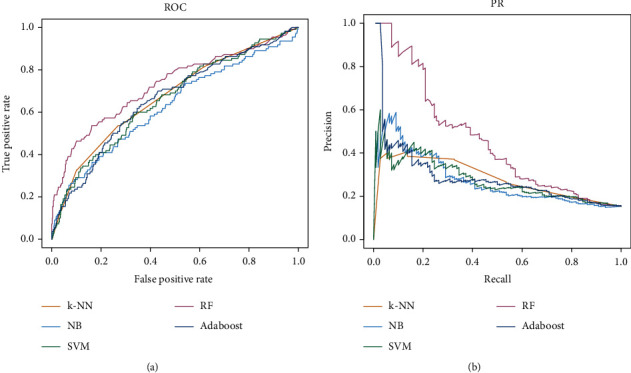
ROC and PR curves of five machine learning models for classification prediction.
Through evaluation and comparison of the predictive ability of the different models, we found that the classification index scores of the random forest model were generally better than those of the other prediction classifiers. Therefore, the random forest model was adopted as the combined drug prediction classifier for this study. The parameter was represented as ntree = 300.
3.5. Prediction Results of Integrative Medicine
We entered “Rheumatoid Arthritis” into the TCMSP database as a search term and identified 41 small molecule types in TCM ingredients that have been used to treat RA. We collected the target treatment information for each small molecule and verified it using the Uniprot database (https://www.uniprot.org/). We collected the molecular formula, PubChem CID, CAS number, and canonical SMILES data of each small molecule from PubChem (Table 3).
Table 3.
Small molecules of 41 anti-RA TCM ingredients collected from TCMSP database.
| TCMSP ID | Name | Molecular formula | PubChem CID | CAS |
|---|---|---|---|---|
| MOL006505 | (-)-Epicatechin | C15H14O6 | 72276 | 490-46-0 |
| MOL006821 | (-)-Epigallocatechin gallate | C22H18O11 | 65064 | 989-51-5 |
| MOL008680 | Acetaldehyde | C2H4O | 177 | 75-07-0 |
| MOL000475 | Anethole | C10H12O | 637563 | 104-46-1 |
| MOL000008 | Apigenin | C15H10O5 | 5280443 | 520-36-5 |
| MOL002773 | Beta-carotene | C40H56 | 5280489 | 7235-40-7 |
| MOL000358 | Beta-Sitosterol | C29H50O | 222284 | 83-46-5 |
| MOL003973 | Caffeine | C8H10N4O2 | 2519 | 58-08-2 |
| MOL008842 | Chenodeoxycholic acid | C24H40O4 | 10133 | 474-25-9 |
| MOL000390 | Daidzein | C15H10O4 | 5281708 | 486-66-8 |
| MOL000254 | Eugenol | C10H12O2 | 3314 | 97-53-0 |
| MOL013179 | Fisetin | C15H10O6 | 5281614 | 528-48-3 |
| MOL000392 | Formononetin | C16H12O4 | 5280378 | 485-72-3 |
| MOL000481 | Genistein | C15H10O5 | 5280961 | 446-72-0 |
| MOL002467 | Gingerol | C17H26O4 | 442793 | 23513-14-6 |
| MOL000666 | Hexanal | C6H12O | 6184 | 66-25-1 |
| MOL005916 | Irisolidone | C17H14O6 | 5281781 | 2345-17-7 |
| MOL000422 | Kaempferol | C15H10O6 | 5280863 | 520-18-3 |
| MOL000305 | Lauric acid | C12H24O2 | 3893 | 143-07-7 |
| MOL000006 | Luteolin | C15H10O6 | 5280445 | 491-70-3 |
| MOL007990 | Militarin | C34H46O17 | 171638 | 58139-23-4 |
| MOL002008 | Myricetin | C15H10O8 | 5281672 | 529-44-2 |
| MOL003493 | Naphthalene | C10H8 | 931 | 91-20-3 |
| MOL003403 | Nicotine | C10H14N2 | 89594 | 54-11-5 |
| MOL005828 | Nobiletin | C21H22O8 | 72344 | 478-01-3 |
| MOL006214 | Progesterone | C21H30O2 | 5994 | 57-83-0 |
| MOL012297 | Puerarin | C21H20O9 | 5281807 | 3681-99-0 |
| MOL000098 | Quercetin | C15H10O7 | 5280343 | 117-39-5 |
| MOL012744 | Resveratrol | C14H12O3 | 445154 | 501-36-0 |
| MOL002268 | Rhein | C15H8O6 | 10168 | 478-43-3 |
| MOL011865 | Rosmarinic acid | C18H16O8 | 5281792 | 20283-92-5 |
| MOL006356 | Sorbitol | C6H14O6 | 5780 | 50-70-4 |
| MOL007154 | Tanshinone IIA | C19H18O3 | 164676 | 568-72-9 |
| MOL003186 | Tripterine | C29H38O4 | 122724 | 34157-83-0 |
| MOL003187 | Triptolide | C20H24O6 | 107985 | 38748-32-2 |
| MOL004932 | Uralsaponin A | C42H62O16 | 128229 | 103000-77-7 |
| MOL000511 | Ursolic acid | C30H48O3 | 64945 | 77-52-1 |
| MOL000635 | Vanillin | C8H8O3 | 1183 | 121-33-5 |
| MOL000173 | Wogonin | C16H12O5 | 5281703 | 632-85-9 |
| MOL009357 | Yakuchinone A | C20H24O3 | 133145 | 78954-23-1 |
| MOL009358 | Yakuchinone B | C20H22O3 | 6440365 | 81840-57-5 |
The information for drugs commonly used in RA treatment was also obtained from DrugBank. In this study, acetaminophen, celecoxib, choline salicylate, diclofenac, hydroxychloroquine, indometacin, leflunomide, methotrexate, naproxen, sulfasalazine, and 10 other common drugs were selected, as presented in Table 4.
Table 4.
Information from DrugBank database for 10 common anti-RA small molecule drugs.
| ID | Name | Molecular formula | PubChem CID | CAS |
|---|---|---|---|---|
| DB00316 | Acetaminophen | C8H9NO2 | 1983 | 103-90-2 |
| DB00328 | Indometacin | C19H16ClNO4 | 3715 | 53-86-1 |
| DB00482 | Celecoxib | C17H14F3N3O2S | 2662 | 169590-42-5 |
| DB00563 | Methotrexate | C20H22N8O5 | 126941 | 59-05-2 |
| DB00586 | Diclofenac | C14H11Cl2NO2 | 3033 | 15307-86-5 |
| DB00788 | Naproxen | C14H14O3 | 156391 | 22204-53-1 |
| DB00795 | Sulfasalazine | C18H14N4O5S | 5339 | 599-79-1 |
| DB01097 | Leflunomide | C12H9F3N2O2 | 3899 | 75706-12-6 |
| DB01611 | Hydroxychloroquine | C18H26ClN3O | 3652 | 118-42-3 |
| DB14006 | Choline salicylate | C12H19NO4 | 54686350 | 2016-36-6 |
The aforementioned 41 small molecule TCM (collected from TCMSP) and 10 common anti-RA Western medicine drugs were combined in pairs to obtain potential drug combinations: C411 × C101 = 410 combinations. Thus, we obtained the final prediction sample set (Supplementary 3; drug combination prediction dataset of integrated Western and TCM drugs).
The results revealed that out of 1000 random sample predictions, 34, 62, and 114 pairs of Western and TCM drugs were predicted to be combination drugs more than 500, 100–500, and 1–100 times, and 200 groups were predicted to be noncombination drugs. In this study, we considered predictions of more than 100 times to be potential anti-RA Western and TCM combination drugs, with higher numbers of positive sample predictions indicating a greater potential for drug combination. Therefore, we selected the 15 groups that were predicted more than 700 times for subsequent cell experiments. The combination drug information is displayed in Table 5 (Supplementary 4; prediction results of all Western and TCM combinations).
Table 5.
Combination data for 15 most predicted drug combinations.
| Group | TCM ingredient | Anti-RA drug | Count |
|---|---|---|---|
| 1 | Eugenol | Acetaminophen | 993 |
| 2 | (-)-Epicatechin | Celecoxib | 957 |
| 3 | Rosmarinic acid | Celecoxib | 946 |
| 4 | Chenodeoxycholic acid | Acetaminophen | 929 |
| 5 | Eugenol | Diclofenac | 912 |
| 6 | Eugenol | Naproxen | 910 |
| 7 | Myricetin | Celecoxib | 908 |
| 8 | Chenodeoxycholic acid | Diclofenac | 898 |
| 9 | Chenodeoxycholic acid | Naproxen | 897 |
| 10 | Rhein | Celecoxib | 894 |
| 11 | Anethole | Naproxen | 854 |
| 12 | Anethole | Diclofenac | 853 |
| 13 | Nobiletin | Celecoxib | 832 |
| 14 | Fisetin | Celecoxib | 791 |
| 15 | Rhein | Hydroxychloroquine | 770 |
3.6. Experimental Verification Results
Because RAW264.7 cells were used for experimental verification, we first determined whether a single small molecule had an inhibitory effect on RAW264.7 cells. We then tested the Western and TCM drug combinations. Finally, the CI value was calculated using CompuSyn software to determine whether the small molecule combinations of the two drugs were synergetic.
We then applied the method described in Section 2.5. Based on observations of the proliferation effect of the 15 small molecules on the RAW264.7 cells, myricetin, nobiletin, fisetin, celecoxib, and hydroxychloroquine significantly reduced the survival rate of the RAW264.7 cells. The IC50 values of the myricetin, nobiletin, fisetin, celecoxib, and hydroxychloroquine were> 100, 23.93, 31.19, >100, and 79.43 μmol/L, respectively, as presented in Figure 6.
Figure 6.
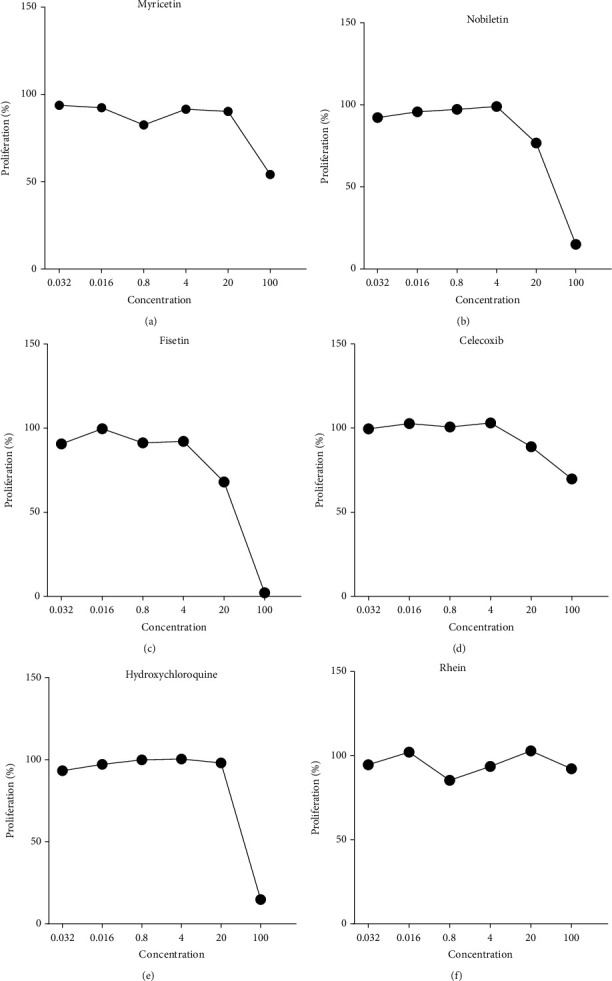
Effects of myricetin, nobiletin, fisetin, celecoxib, hydroxychloroquine, and rhein on the viability of RAW264.7 cells.
We further tested combinations of Western and TCM containing these five small molecules (a total of seven pairs) based on the prediction results of the machine learning model and calculated the CI values, as displayed in Table 6. We found that of the seven pairs of Western and TCM combinations, five of the pairs had CI values less than 0.7, indicating that these drugs have good synergy.
Table 6.
Effect of seven predicted pairs of Western and TCM combinations on RAW264.7 cell viability.
| Combination | TCM (μmol/L) | Drug (μmol/L) | IC50 | RAW264.7 cell inhibition rate (%) | CI |
|---|---|---|---|---|---|
| Rhein+celecoxib | >100 + 100 | CI < 1 | |||
| 100 | 100 | 39.31 ± 2.92 | CI = 0.37 | ||
| 20 | 100 | 35.68 ± 1.49 | CI = 0.49 | ||
| 4 | 100 | 38.60 ± 0.80 | CI = 0.39 | ||
| 100 | 20 | 20.08 ± 1.22 | CI = 0.40 | ||
| 100 | 4 | 14.22 ± 1.59 | CI = 0.17 | ||
| 20 | 20 | 20.17 ± 1.49 | CI = 0.40 | ||
|
| |||||
| Nobiletin+celecoxib | 8.83 + 100 | CI < 1 | |||
| 100 | 100 | 96.20 ± 0.24 | CI = 0.01 | ||
| 100 | 20 | 61.50 ± 1.67 | CI = 0.17 | ||
|
| |||||
| Myricetin+celecoxib | 100 + 4 | CI < 1 | |||
| 100 | 100 | 64.05 ± 0.94 | CI = 0.06 | ||
| 20 | 100 | 37.13 ± 2.13 | CI = 0.46 | ||
| 4 | 100 | 37.96 ± 1.58 | CI = 0.41 | ||
| 100 | 20 | 53.92 ± 1.59 | CI = 0.03 | ||
| 100 | 4 | 49.97 ± 0.93 | CI = 0.02 | ||
|
| |||||
| Fisetin+celecoxib | 10.89 + 100 | CI < 1 | |||
| 100 | 100 | 99.06 ± 0.03 | CI = 0.02 | ||
| 4 | 100 | 57.80 ± 1.89 | CI = 0.88 | ||
|
| |||||
| Rhein+hydroxychloroquine | 100 + 57.15 | CI < 1 | |||
| 100 | 20 | 86.88 ± 0.04 | CI = 0.28 | ||
|
| |||||
| Epicatechin+celecoxib | >100 + 100 | CI > 1 | |||
|
| |||||
| Rosmarinic acid+celecoxib | >100 + 100 | CI > 1 | |||
4. Discussion
Machine learning methods have been widely used in many fields of research, such as text classification [23], sentiment analysis [24], breast cancer diagnosis [25], web page classification [26], and image generation [27]. Many researchers have used computational and modeling methods to identify potential combinatorial drugs [17, 28–30], and the use of machine learning to predict drug combinations has notably increased in recent years [31–34]. However, nearly all of this research has focused of combinations of Western drugs. Combinations of Western and TCM drugs have rarely been reported, especially within the context of RA treatment. TCM is a complex system composed of many small molecules with synergistic effects. In recent years, the research of TCM small molecule is still the focus of modern pharmacology. We mainly focused on the prediction of the combination of small molecule with clear pharmacological effects in TCM and Western medicines that have been approved drugs. Therefore, in this study, we examined the small molecule characteristics of TCM ingredients that differed from those in Western combination drugs. We developed a method for complete modeling and prediction to screen anti-RA combination drugs.
For feature variable construction, we selected drug structures and ADME features. However, the results of the statistical analyses and feature variable screening revealed that drug structure similarity and ADME characteristics had no significant effect on the classification of combination drugs. Notably, sequence similarity, GO functional similarity, and the distance of drug targets within the PPI network of the small molecule drug targets were found to be related to the combination of drugs, with the similarity value of the drug target pathway (SPathway) being identified as a key feature in combination drugs, both statistically and in feature screening. Therefore, the synergistic mechanism of two or more drugs within a pathway may serve as a key reference for future developments of combined drugs.
In constructing the sample training set, we were unable to collect sufficient noncombined drug data (namely the number of negative samples) due to limited research data. Therefore, in the original data, the number of positive samples was only 1% that of negative samples, which created a serious imbalance in the number of positive and negative samples. If the model had been constructed using the original datasets, the disproportionate number of negative samples would have caused overfitting, and the predictions would have been biased toward the classifications of the larger number of samples. This would have greatly reduced the normalizing ability of the models. However, simply reducing the sample size may have excluded potential drug combinations in the 45603 samples.
To effectively and reasonably overcome this challenge, we proposed a method in which multiple training sets were constructed using random sampling performed 1000 times. For each instance of modeling, the positive samples remained unchanged, and a limited number of negative samples were randomly selected from the total negative samples; 1000 training sets were randomly composed. The machine learning model was trained by and predicted the 1000 training sets. The final potential combination drugs were then determined on the basis of the percentage of each sample predicted to be a combination drug (positive) in the 1000 learning sessions. However, because the number of noncombination drugs is generally much larger than that of combination drugs but large deviations in numbers of positive and negative samples would cause prediction deviation in the machine learning model, in this study, the ratio of positive and negative samples was set to 1 : 5. Through these changes, we were able to prevent imperfect data sampling of combined drugs.
In this study, from the prediction and evaluation results of five different machine learning classification models in 1000 random sample training sets, the accuracy, precision, F1 score, AUROC, and AUPR values of RF are the highest among the five machine learning models. AdaBoost performs best on recall, followed by SVM and RF. The prediction scores of k-NN and NB on these six machine learning evaluation indexes were lower, suggesting that these two models may not be suitable for combination drug prediction. The prediction performance of SVM and AdaBoost is second only to that of RF. Therefore, through the evaluation and comparison of the predictive ability of different models, we found that the RF classification index scores are generally better than other predictive classifiers. Therefore, the RF model is used as the final combined drug prediction classifier, where the parameter ntree = 300. We used the RF classifier to predict 410 groups of Western and TCM drug combinations to treat RA and thus preliminarily predicted 96 pairs of potential Western and TCM drug combinations to treat RA. We used RAW264.7 cells to experimentally verify the effects of a single drug small molecule on the proliferation of RAW264.7 cells. We then verified the effects of combined drugs on the cells. The results revealed that five of the seven selected pairs of combined Western and TCM drugs had high synergistic effects, indicating that our model had high accuracy and practicability. The five optimal combinations were Rhein and Celecoxib, Nobiletin and Celecoxib, Myricetin and Celecoxib, Fisetin and Celecoxib, and Rhein and Hydroxychloroquine.
Notably, celecoxib was found to have a higher synergistic therapeutic effect when combined with several small molecule TCM ingredients, namely, myricetin, rhein, nobiletin, and fisetin. In addition, rhein did not demonstrate an inhibitory effect on RAW264.7 cell proliferation when applied alone. However, when combined with celecoxib (CI < 0.7), the combined inhibition rate of 100 μmol/L rhein and 20 μmol/L celecoxib was 35.68%, which was higher than the rate of each of the two alone. Rhien had high synergy with hydroxychloroquine.
Notably, triptolide combined with Western drugs, such as methotrexate, has previously been reported to produce favorable results in past experiments [35]. However, the number of predicted combinations containing triptolide in our model was not high; it predicted triptolide combined with diclofenac (75 times), naproxen (75 times), and celecoxib (24 times) but did not predict triptolide combined with methotrexate (0 times). This may be due to triptolide's reported clinical toxicity [36]. Although triptolide has been clinically reported to improve treatment outcomes, it may not be the best option for drug combinations and may even lead to more systemic side effects [37]. Our prediction results may support this hypothesis. The screening of potential combination of TCM and Western medicine based on the machine learning combined with experimental validation methods can improve efficiency and reduce research costs.
In conclusion, we constructed combination drug characteristics based on existing combination drug data and characteristics from TCM drug research. Key characteristic indexes were screened, and a random forest classification model was used to predict potential combinations of Western and TCM drugs for anti-RA treatment. Through experimental verification, we preliminarily demonstrated that this method of combination therapy for anti-RA treatment can be applied to clinical practice. In the future, we will conduct vivo experiments and clinical trials to validate the findings of this study. In addition, our machine learning model for integrated Western and TCM drug combination can be applied for predicting not only anti-RA drug combinations but also TCM combinations for other diseases.
Acknowledgments
This work was supported by the Chinese Medicine Research Project Plan of Shanghai Municipal Health Commission (grant no. 2020JP002), Shanghai Natural Science Fund (grant no. 19ZR1452000), Shanghai Municipality which further accelerates the Three-Year Action Plan for the Development of Traditional Chinese Medicine (2018–2020), the “Internet + TCM Health Service” Research and Transformation Platform Construction Fund (grant no. ZY (2018-2020)-CCCX-2001-01), and Shanghai Collaborative Innovation Center for Chronic Disease Prevention and Health Services (2021 Science and Technology 02-37).
Abbreviations
- ADME:
Adsorption, distribution, metabolism, and excretion
- AUPRC:
Area under the precision–recall curve
- CAS:
Chemical Abstracts Service
- CI:
Combination index
- DMARDs:
Disease modifying antirheumatic drugs
- FDA:
Food and Drug Administration
- FN:
False negative
- FP:
False positive
- FPR:
False positive rate
- GO:
Gene ontology
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- k-NN:
k-nearest neighbors
- NB:
Naive Bayes
- NSAIDs:
Nonsteroidal anti-inflammatory drugs
- PPI:
Protein–protein interaction
- RA:
Rheumatoid arthritis
- RF:
Random forest
- ROC:
Receiver operating characteristic
- SMILES:
Canonical simplified molecular-input line-entry system
- SVM:
Support vector machine
- TCM:
Traditional Chinese medicine
- TCMSP:
Traditional Chinese Medicine Systems Pharmacology
- TN:
True negative
- TP:
True positive
- TPR:
True positive rate.
Contributor Information
Jianying Wang, Email: wjy8310@163.com.
Lei Zhang, Email: zhanglei37@sina.com.
Jihan Huang, Email: huangjihan@shutcm.edu.cn.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors have no conflicts of interest to declare regarding the publication of this article.
Supplementary Materials
Pretreated FDA-certified combination drug dataset.
Positive and negative sample training dataset for all drug combinations.
Drug combination prediction dataset of integrated Western and TCM drugs.
Prediction results of all Western and TCM combinations.
References
- 1.Chou T. C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacological Reviews . 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 2.Bokhari S. U., Gopal U. M., Duckworth W. C. Beneficial effects of a glyburide/metformin combination preparation in type 2 diabetes mellitus. The American Journal of the Medical Sciences . 2003;325(2):66–69. doi: 10.1097/00000441-200302000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Augustin K., Williams S., Cunningham M., et al. Perampanel and decanoic acid show synergistic action against AMPA receptors and seizures. Epilepsia . 2018;59(11):e172–e178. doi: 10.1111/epi.14578. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z. Y., Chen M., Chen J., Padval M. V., Kansra V. V. Biotransformation and in vitro assessment of metabolism-associated drug-drug interaction for CRx-102, a novel combination drug candidate. Journal of Pharmaceutical and Biomedical Analysis . 2009;50(2):200–209. doi: 10.1016/j.jpba.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhari K., Rizvi S., Syed B. A. Rheumatoid arthritis: current and future trends. Nature Reviews. Drug Discovery . 2016;15(5):305–306. doi: 10.1038/nrd.2016.21. [DOI] [PubMed] [Google Scholar]
- 6.Walsh D. A., McWilliams D. F. Mechanisms, impact and management of pain in rheumatoid arthritis. Nature Reviews Rheumatology . 2014;10(10):581–592. doi: 10.1038/nrrheum.2014.64. [DOI] [PubMed] [Google Scholar]
- 7.Chung I. M., Ketharnathan S., Thiruvengadam M., Rajakumar G. Rheumatoid arthritis: the stride from research to clinical practice. International Journal of Molecular Sciences . 2016;17(6):p. 900. doi: 10.3390/ijms17060900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gossec L., Dougados M. Combination therapy in early rheumatoid arthritis. Clinical and Experimental Rheumatology . 2003;21(5) Supplement 31:S174–S178. [PubMed] [Google Scholar]
- 9.Hazlewood G. S., Barnabe C., Tomlinson G., et al. Methotrexate monotherapy and methotrexate combination therapy with traditional and biologic disease modifying anti-rheumatic drugs for rheumatoid arthritis: a network meta-analysis. Cochrane Database of Systematic Reviews . 2016;2016(8, article Cd010227) doi: 10.1002/14651858.CD010227.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia Z., Lyu J., Hou N., Song L., Li X., Liu H. Iguratimod in combination with methotrexate in active rheumatoid arthritis. Zeitschrift für Rheumatologie . 2016;75(8):828–833. doi: 10.1007/s00393-015-1641-y. [DOI] [PubMed] [Google Scholar]
- 11.Ibfelt E. H., Jacobsen R. K., Kopp T. I., et al. Methotrexate and risk of interstitial lung disease and respiratory failure in rheumatoid arthritis: a nationwide population-based study. Rheumatology . 2021;60(1):346–352. doi: 10.1093/rheumatology/keaa327. [DOI] [PubMed] [Google Scholar]
- 12.Albrecht K., Müller-Ladner U. Side effects and management of side effects of methotrexate in rheumatoid arthritis. Clinical and Experimental Rheumatology . 2010;28(5) Supplement 61:S95–101. [PubMed] [Google Scholar]
- 13.Wang W., Wang X., Tang X., Jiang Q., Fan Y. Classifying rheumatoid arthritis by traditional Chinese medicine Zheng: a multi-center cross-sectional study. Journal of Traditional Chinese Medicine . 2019;39(3):425–432. [PubMed] [Google Scholar]
- 14.Zhang P., Li J., Han Y., Yu X. W., Qin L. Traditional Chinese medicine in the treatment of rheumatoid arthritis: a general review. Rheumatology International . 2010;30(6):713–718. doi: 10.1007/s00296-010-1370-0. [DOI] [PubMed] [Google Scholar]
- 15.Liu H., Zhang W., Zou B., Wang J., Deng Y., Deng L. DrugCombDB: a comprehensive database of drug combinations toward the discovery of combinatorial therapy. Nucleic Acids Research . 2020;48(D1):D871–D881. doi: 10.1093/nar/gkz1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports . 2017;7(1, article 42717) doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng F., Kovács I. A., Barabási A. L. Network-based prediction of drug combinations. Nature Communications . 2019;10(1, article 1197) doi: 10.1038/s41467-019-09186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cover T., Hart P. Nearest neighbor pattern classification. IEEE Transactions on Information Theory . 1967;13(1):21–27. doi: 10.1109/TIT.1967.1053964. [DOI] [Google Scholar]
- 19.Kohonen J., Talikota S., Corander J., Auvinen P., Arjas E. A naive Bayes classifier for protein function prediction. In Silico Biology . 2009;9(1,2):23–34. doi: 10.3233/ISB-2009-0382. [DOI] [PubMed] [Google Scholar]
- 20.Zhang T. An introduction to support vector machines and other kernel-based learning methods. AI Magazine . 2001;30:p. 103. [Google Scholar]
- 21.Pal M. Random forest classifier for remote sensing classification. International Journal of Remote Sensing . 2007;26:217–222. doi: 10.1080/01431160412331269698. [DOI] [Google Scholar]
- 22.Freund Y., Schapire R. E. A short introduction to boosting. Journal of Japanese Society for Artificial Intelligence . 1999;14, article 1612 [Google Scholar]
- 23.Onan A., Korukoğlu S., Bulut H. Ensemble of keyword extraction methods and classifiers in text classification. Expert Systems with Applications . 2016;57:232–247. doi: 10.1016/j.eswa.2016.03.045. [DOI] [Google Scholar]
- 24.Onan A. Sentiment analysis on product reviews based on weighted word embeddings and deep neural networks. Concurrency and Computation Practice and Experience . 2020;33(23, article e5909) doi: 10.1002/cpe.5909. [DOI] [Google Scholar]
- 25.Onan A. A fuzzy-rough nearest neighbor classifier combined with consistency-based subset evaluation and instance selection for automated diagnosis of breast cancer. Expert Systems with Applications . 2015;42(20):6844–6852. doi: 10.1016/j.eswa.2015.05.006. [DOI] [Google Scholar]
- 26.Onan A. Classifier and feature set ensembles for web page classification. Journal of Information Science . 2016;42(2):150–165. doi: 10.1177/0165551515591724. [DOI] [Google Scholar]
- 27.Abazari M. A., Soltani M., Moradi Kashkooli F., Raahemifar K. Synthetic 18F-FDG PET image generation using a combination of biomathematical modeling and machine learning. Cancers . 2022;14(11):p. 2786. doi: 10.3390/cancers14112786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao X. M., Iskar M., Zeller G., Kuhn M., van Noort V., Bork P. Prediction of drug combinations by integrating molecular and pharmacological data. PLoS Computational Biology . 2011;7(12, article e1002323) doi: 10.1371/journal.pcbi.1002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y. Y., Xu K. J., Song J., Zhao X. M. Exploring drug combinations in genetic interaction network. BMC Bioinformatics . 2012;13(Supplement 7):p. S7. doi: 10.1186/1471-2105-13-S7-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H., Zhang P., Qu X. A., Sanseau P., Yang L. Systematic prediction of drug combinations based on clinical side-effects. Scientific Reports . 2014;4(1, article 7160) doi: 10.1038/srep07160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y., Xiong Y., Xu Q., Wei D. A hadoop-based method to predict potential effective drug combination. BioMed Research International . 2014;2014:5. doi: 10.1155/2014/196858.196858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Qin G., Yang Q., Chen L., Xie L. Biomolecular network-based synergistic drug combination discovery. BioMed Research International . 2016;2016:11. doi: 10.1155/2016/8518945.8518945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X., Ren B., Chen M., Wang Q., Zhang L., Yan G. NLLSS: predicting synergistic drug combinations based on semi-supervised learning. PLoS Computational Biology . 2016;12(7, article e1004975) doi: 10.1371/journal.pcbi.1004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabben H. L., Andersen G. T., Ianevski A., et al. Computational drug repositioning and experimental validation of ivermectin in treatment of gastric cancer. Frontiers in Pharmacology . 2021;12, article 625991 doi: 10.3389/fphar.2021.625991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W., Gong K., Ding Y., et al. Effects of triptolide and methotrexate nanosuspensions on left ventricular remodeling in autoimmune myocarditis rats. International Journal of Nanomedicine . 2019;14:851–863. doi: 10.2147/IJN.S191267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xi C., Peng S., Wu Z., Zhou Q., Zhou J. Toxicity of triptolide and the molecular mechanisms involved. Biomedicine & Pharmacotherapy . 2017;90:531–541. doi: 10.1016/j.biopha.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Grzegorzewska A. E., Frankiewicz D., Bręborowicz D., Matławska I., Bylka W. Disseminated cutaneous Kaposi sarcoma in a patient receiving triptolide/tripdiolide for rheumatoid arthritis. Medical Science Monitor . 2012;18(8, article Cs67) doi: 10.12659/MSM.883256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pretreated FDA-certified combination drug dataset.
Positive and negative sample training dataset for all drug combinations.
Drug combination prediction dataset of integrated Western and TCM drugs.
Prediction results of all Western and TCM combinations.
Data Availability Statement
The data used to support the findings of this study are included within the article.


