Abstract
A d-xylulose 5-phosphate/d-fructose 6-phosphate phosphoketolase (Xfp) from the probiotic Bifidobacterium lactis was purified to homogeneity. The specific activity of the purified enzyme with d-fructose 6-phosphate as a substrate is 4.28 Units per mg of enzyme. Km values for d-xylulose 5-phosphate and d-fructose 6-phosphate are 45 and 10 mM, respectively. The native enzyme has a molecular mass of 550,000 Da. The subunit size upon sodium dodecyl sulfate-polyacrylamide gel electrophoresis (90,000 Da) corresponds with the size (92,529 Da) calculated from the amino acid sequence of the isolated gene (named xfp) encoding 825 amino acids. The xfp gene was identified on the chromosome of B. lactis with the help of degenerated nucleotide probes deduced from the common N-terminal amino acid sequence of both the native and denatured enzyme. Comparison of the deduced amino acid sequence of the cloned gene with sequences in public databases revealed high homologies with hypothetical proteins (26 to 55% identity) in 20 microbial genomes. The amino acid sequence derived from the xfp gene contains typical thiamine diphosphate (ThDP) binding sites reported for other ThDP-dependent enzymes. Two truncated putative genes, pta and guaA, were localized adjacent to xfp on the B. lactis chromosome coding for a phosphotransacetylase and a guanosine monophosphate synthetase homologous to products of genes in Mycobacterium tuberculosis. However, xfp is transcribed in B. lactis as a monocistronic operon. It is the first reported and sequenced gene of a phosphoketolase.
Phosphoketolases (EC 4.1.2.9, EC 4.1.2.22) are thiamine diphosphate (ThDP)-dependent key enzymes of the pentose phosphate pathway of heterofermentative and facultative homofermentative lactic acid bacteria and of the d-fructose 6-phosphate (F6P) shunt of bifidobacteria (1, 2, 9, 13, 15, 29, 32). Phosphoketolases have been sporadically reported in other microorganisms (11, 14, 30). In bifidobacteria, two types of phosphoketolases have been described: a F6P specific enzyme (F6PPK) in human species like B. dentium, and a dual substrate xylulose 5-phosphate/fructose 6-phosphate (X5P/F6P) phosphoketolase (Xfp) in animal species like B. globosum (12, 31). No molecular data are available for any of the phosphoketolases defining for one active protein molecular size, subunit size, and amino acid sequence. As a preliminary to studies of the biochemistry, genetics, and regulation of these landmark enzymes in the nutritionally important bifidobacteria of the human and animal intestine, we report the purification of the dual substrate Xfp from Bifidobacterium lactis, a bacterium used as a probiotic supplement in fermented food (16, 22). The gene (xfp) was identified, cloned, and sequenced.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bifidobacterium lactis was grown anaerobically at 37°C in Bif medium (brain heart infusion [37 g/liter; Biolife] supplemented with yeast extract [5 g/liter], l-cysteine hydrochloride [0.5 g/liter], and resazurine [2 mg/liter]) as described before (22). Escherichia coli was routinely grown at 37°C in Luria-Bertani medium (27). If required, the following supplements were added to the media: ampicillin, 50 μg/ml; spectinomycin, 50 μg/ml; tetracycline, 10 μg/ml; kanamycin, 30 μg/ml; chloramphenicol, 30 μg/ml; 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 20 μg/ml; and isopropylthio-β-d-galactopyranoside (IPTG), 240 μg/ml.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristics, construction, or nucleotide sequence (5′ → 3′)a | Reference or source |
|---|---|---|
| Strains | ||
| B. lactis DSM 10140 | Wild type | 22 |
| E. coli XL1-Blue | recA1 lac endA1 gyrA96 thi hsdR17 supE44 relA1 (F′ proAB lacIq lacZΔM15 Tn10 Tetr) | 3 |
| E. coli BL21(DE3)/pLysS | F−ompT hsdSB(rB− mB−) gal dcm (DE3)/pLysS (Cmr) | Novagen |
| Plasmids | ||
| pUC18 | Ampr LacZ′; cloning vector; 2.7 kb | 35 |
| pCL1920 | Spcr Strr; cloning vector; 4.6 kb | 19 |
| pGEM-T Easy | Ampr LacZ′; annealed T in both 3′ ends after linearization with EcoRV; 3.0 kb | Promega |
| pET-28a(+) | Kanr LacI; expression vector; 5.4 kb | Novagen |
| pFPK1 | Ampr; 2.62-kb BamHI-BamHI fragment from strain DSM 10140 in pUC18; 5.3 kb | This study |
| pFPK2 | Ampr; 1.6-kb PCR-derived fragment from strain DSM 10140 in pGEM-T Easy; 4.6 kb | This study |
| pFPK3 | Spcr Strr; 0.85-kb BamHI-SmaI fragment from pFPK2 insertion in pCL1920; 5.4 kb | This study |
| pFPK4 | Spcr Strr; 2.62-kb BamHI-insertion from pFPK1 in the BamHI site of pFPK3; 8.1 kb | This study |
| pFPK5 | Kanr; 2.5-kb PCR-derived fragment from strain DSM 10140 in pET-28a(+); 7.9 kb | This study |
| Oligonucleotidesb | ||
| pk5 | 5′-GGIACICCITGGCARAAR-3′ (974–991) | This study |
| pk6 | 5′-ATATATATARTAYTTRTCCATICCIATIAT-3′ (1019–1039 reverse)c | This study |
| pk7 | 5′-CATGGCAGAAGCTGGATCGTCCGGT-3′ (981–1005) | This study |
| pk9 | 5′-GCGAGATCCCGTGGCGT-3′ (2526–2542) | This study |
| pk15 | 5′-ATATATGAATTCATGACTAATCCTGTTATTGGTACC-3′ (956–979)c | This study |
| pk16 | 5′-AATTACAAGCTTTCACTCGTTGTCGCCGGCGG-3′ (3414–3433 reverse)c | This study |
Position in the nucleotide sequence according to the numbering in the GenBank database.
Oligonucleotides were synthesized by Microsynth AG, Balgach, Switzerland.
The underlined sequences correspond to an introduced tail for cloning purposes.
Preparation of protein extracts.
All procedures were carried out at 0 to 4°C. B. lactis cells were harvested from liquid medium in the exponential growth phase (A600 between 1.2 and 1.4) by centrifugation (10,000 × g for 20 min), washed twice with a saline solution (0.85 M NaCl) and resuspended in 4 ml/g (wet weight) of buffer A, composed of 100 mM potassium phosphate containing 30 mM KCl, 0.1 mM EDTA, 1 mM MgCl2, 0.2 mM phenylmethylsulfonyl fluoride, and 2 mM 1,4-dithio-dl-threitol at pH 7.0. Cells were disrupted by 12 passages through a chilled French pressure cell (SLM Amico; Lightening Instruments, Lausanne, Switzerland) at approximately 120 MPa. After centrifugation at 20,000 × g for 40 min, the clarified supernatant was extensively dialyzed against buffer A to get a so-called crude extract. Crude extracts of E. coli recombinant clones were obtained by the same procedure with the exception that only three passages through the French pressure cell were performed.
Purification of phosphoketolase activity.
Column chromatographies were done at room temperature. All other procedures in between were performed at 4°C. The amount of protein used in a typical purification are given in Table 2.
TABLE 2.
Purification of Xfp from B. lactis
| Step | Vol (ml) | Total units | Total protein (mg) | Sp act (Ua/mg) | Purification factor | Yield (%) |
|---|---|---|---|---|---|---|
| Crude extract | 15 | 93.4 | 246.0 | 0.38 | 1.0 | 100 |
| DEAE-cellulose | 138 | 28.7 | 47.1 | 0.61 | 1.6 | 30.7 |
| Mono Q | 2.1 | 15.5 | 7.4 | 2.10 | 5.5 | 16.6 |
| Superdex 200 | 6.0 | 4.1 | 0.96 | 4.28 | 11.1 | 4.4 |
One unit is defined as the amount of extract forming 1 μmol of acetyl phosphate per min with F6P as a substrate.
(i) Step 1: DEAE-anion-exchange chromatography.
The crude extract was loaded onto a column (15 by 2.6 cm) of DEAE Sepharose Fast Flow (Pharmacia) equilibrated with buffer A. Proteins were eluted in four subsequent steps with buffer A at potassium phosphate concentrations of 100, 150, 300, and 600 mM, respectively, and at a flow rate of 1.1 ml/min (corresponding to 12.5 cm/h). In each step, the column was eluted with 4.4 column volumes of the corresponding buffer. The fractions containing F6PPK activity which eluted at a phosphate buffer concentration of 300 mM were pooled and concentrated by membrane filtration (Centriprep YM-10, 10,000 Da; Millipore). The concentrated sample was dialyzed overnight against 20 mM Tris-HCl at pH 8.0 (buffer B).
(ii) Step 2: Mono Q-anion-exchange chromatography.
The resulting sample (2.1 ml) was applied to a 1-ml Mono Q 5/5 anion-exchange fast-performance liquid chromatography column (Pharmacia) which had been equilibrated with buffer B. After a 10-ml wash, the column was eluted with a 90-ml linear gradient from 0 to 500 mM KCl in buffer B. The fractions containing F6PPK activity (peak fraction at approximately 450 mM KCl) were pooled and concentrated by membrane filtration (10,000 Da).
(iii) Step 3: size exclusion chromatography.
After a buffer exchange by dialysis against buffer C (Tris-HCl at pH 7.5 containing 150 mM NaCl), the sample from step 2 was subjected to gel filtration chromatography with a fast-performance liquid chromatography Superdex 200 HR 10/30 column (Pharmacia) of a volume of 24 ml. Proteins were eluted at a flow rate of 0.5 ml/min. The pooled fractions containing F6PPK activity were stored at −20°C until analytical measurements were carried out. The column had been calibrated with protein molecular weight markers (Bio-Rad Laboratories) thyroglobulin (669,000), ferritin (440,000), catalase (232,000), aldolase (158,000), and albumin (67,000). It was also used to estimate the molecular size of the purified native enzyme.
Phosphoketolase assay and activity unit definition.
Phosphoketolase activity was measured spectrophotometrically as ferric acetyl hydroxamate produced from the enzymatically generated acetyl phosphate by the procedure of Racker (26). The standard reaction mixture of 0.075 ml consisted of 33.3 mM potassium phosphate (pH 6.5), l-cysteine hydrochloride (1.9 mM), sodium fluoride (23 mM), sodium iodoacetate (8 mM), either d-fructose 6-phosphate or d-xylulose 5-phosphate (Fluka) as a substrate (each at a concentration of 27 mM), and finally the protein sample to initiate the reaction. After incubating at 37°C for 30 min, 0.075 ml of hydroxylamine hydrochloride (2 M, pH 6.5) was added at room temperature. Ten minutes later, 0.05 ml of 15% (wt/vol) trichloroacetic acid, 0.05 ml of 4 M HCl, and 0.05 ml of FeCl3 · 6 H2O (5% [wt/vol] in 0.1 M HCl) were added for the final color development of the ferric hydroxamate, which was then spectrophotometrically quantified at 505 nm by comparing to a series of acetyl phosphate (Sigma) standards. For qualitative measurements of F6PPK activity in whole cells, E. coli cells containing recombinant phosphoketolase or B. lactis cells were pretreated with hexadecyltrimethylammonium bromide (Sigma) before being assayed according to the method of Orban and Patterson (24).
One unit of phosphoketolase activity is defined as the amount of extract forming 1 μmol of acetyl phosphate per min from either F6P or X5P. Specific activity is expressed as units per milligram of protein. Protein concentrations were determined by the method of Lowry et al. (20) using bovine serum albumin as a standard.
Protein gel electrophoresis and amino acid analysis.
Protein extracts were analyzed by polyacrylamide gel electrophoresis (PAGE) by established methods (6, 33) and N-terminal amino acids were determined as described elsewhere (18).
DNA isolation and manipulations.
Total cellular DNA was prepared from B. lactis as described by Leenhouts et al. (17) with some modifications: 1.5 g (wt/vol) of frozen B. lactis cells (wet weight) were dissolved in 5 ml of lysis solution (0.025 M Tris-HCl [pH 8.0], 0.05 M EDTA, 0.05 M d-glucose) containing 25 mg of lysozyme (Sigma), 600 U of mutanolysin (Sigma), and 0.1 mg of RNase A (Sigma). This mixture was incubated for 30 min at 37°C and subsequently treated with proteinase K, and the DNA was precipitated as described in the original procedure (17). Electroporation of E. coli cells (8), small-scale plasmid DNA isolation from E. coli, DNA modifications, and further manipulations were carried out by standard methods (27). Transfer of DNA from 0.8% agarose gels to Nylon Plus membrane (QIAGEN) was performed according to the method of Southern (27). Synthetic oligonucleotides (approximately 5 pmol) were end-labeled by phosphorylation with T4 polynucleotide kinase and 0.1 mCi of [γ-32P]ATP (5 × 103 Ci/mmol) (27) whereas plasmid DNA (approximately 0.2 μg) was nick labeled with Klenow polymerase using 0.03 mCi of [α-32P]ATP (3 × 103 Ci/mmol) (27). The conditions for prehybridizations and hybridizations with oligonucleotides or DNA probes were carried out in the absence of formamide as previously described (21).
Cloning the B. lactis phosphoketolase gene.
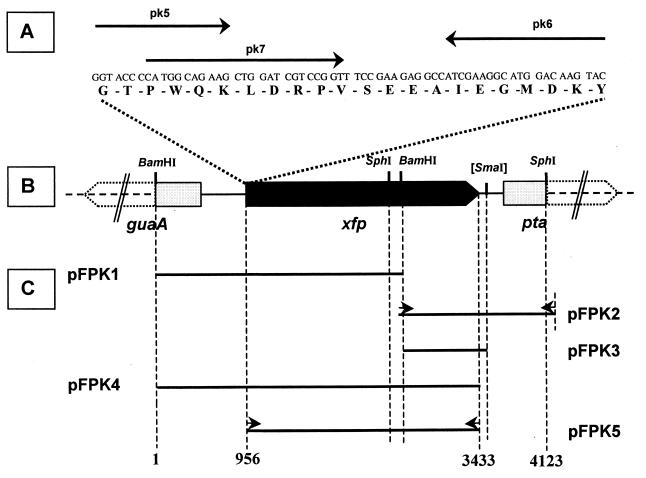
In order to obtain a (short) DNA fragment by PCR, degenerated oligonucleotide primers pk5 and pk6 (Table 1; Fig. 1) were deduced from the N terminus which had been determined for the B. lactis protein catalyzing the phosphoketolase reaction. These primers were used in a “touchdown PCR” using B. lactis DNA as a template. Thereby, the annealing temperature was decreased gradually from 50 to 40°C within the first 20 PCR cycles before holding it at 40°C for the next 15 cycles. Annealing time was 0.5 min. Extension occurred with Taq DNA polymerase (Amersham Pharmacia Biotech) at 72°C for 1 min and DNA strand separation was performed at 95°C for 0.5 min. The resulting PCR product was sequenced and the internal oligonucleotide pk7 was deduced which served as a probe in hybridizations with B. lactis DNA. A hybridizing 2.62-kb BamHI fragment from B. lactis was cloned (pFPK1) (Table 1) and sequenced. An overlapping fragment from the B. lactis chromosome was cloned in a two-step protocol. Step 1 was a ligation of SphI-digested B. lactis DNA (sized to 1.5 to 2.5 kb fragments) followed by transformation of E. coli XL-1 Blue cells and immediate growth in liquid Luria-Bertani medium. In step 2, DNA was extracted from such an overnight culture and then used for a PCR. For that, pk9 targeting the insertion of pFPK1 between the SphI and BamHI site (Fig. 1) and pUC18/M13(−20) (Promega) served as primers at an annealing temperature of 65°C for 0.5 min. Otherwise, the PCR conditions were the same as described above. The resulting PCR product was restricted and cloned into pGEM-T Easy to form plasmid pFPK2 (Table 1) containing a 1.62-kb recombinant fragment. In order to compose the insertions of pFPK1 and pFPK2 in the chromosomal order, an internal 0.85-kb BamHI-SmaI fragment from the pFPK2 insertion was cloned into plasmid pCL1920 to obtain pFPK3 (Table 1 and Fig. 1). Its single BamHI cleavage site was used to finally insert the 2.62-kb BamHI fragment of pFPK1. The resulting plasmid, pFPK4, is supposed to contain the hypothetical phosphoketolase gene as it occurs on the B. lactis chromosome. After sequence analysis, the entire gene was amplified by PCR using B. lactis DNA, primer pair pk15-pk16 (Table 1), and Pfu DNA polymerase (Stratagene), and the resulting PCR product finally cloned into the expression vector pET-28a(+) (Table 1) to form plasmid pFPK5.
FIG. 1.
Cloning strategy and restriction map of the B. lactis xfp gene and its adjacent region. (A) Amino acids (7 to 28) from the N terminus of the Xfp protein, position of deduced primers (arrows), and corresponding nucleotide sequence. (B) Open reading frames guaA, xfp, and pta (open and filled arrows) and restriction sites which were relevant for cloning. (C) Inserts of recombinant plasmids (solid lines) bordered by dashed vertical lines representing either restriction sites or 5′ ends of primers used for PCR; primers are indicated by short arrows and described in detail in Table 1. Numbers at the end of vertical dashed lines are assigned to nucleotide numbers as indicated in the text and in the GenBank database.
Nucleotide sequence determination.
Nucleotide sequencing of both strands from cloned DNA was performed by the dideoxy chain termination method (28) with primer walking using the BigDye Terminator Cycle Sequencing Ready Reaction Kit and the ABI PRISM ABI 310 Genetic Analyzer apparatus (Applied Biosystems, Foster City, Calif.) for analysis. DNA sequence analysis, sequence alignments, and sequence database searching were conducted with programs contained within the Sequence Analysis Software Package (version 10.0) licensed from the Genetics Computer Group (University of Wisconsin, Madison). Sequences were compared by the algorithm of Pearson and Lipman (25) (FastA and TFastA) with sequences in the GenEMBL database copy.
Northern blot hybridization analysis.
For RNA analysis experiments B. lactis was cultivated in 100 ml of Bif medium. RNA from both exponentially growing cells and cells from early stationary growth phase were prepared according to the hot acid-phenol extraction method of Oelmüller et al. (23). After denaturation with formamide, RNA (approximately 5 to 10 μg) was analyzed by gel electrophoresis on agarose containing 20 mM guanidine thiocyanate, followed by electroblotting onto GeneScreen Plus (NEN Life Sciences, Albany, N.Y.) membranes. Hybridization was carried out with [α-32P]ATP-labeled DNA probes in a stringent sodium dodecyl sulfate (SDS) buffer (5) at 65°C.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in GenBank under accession number AJ293946. All nucleotide numbers used in the current study refer on the numbering of the GenBank entry dated 6 September 2000.
RESULTS
Isolation and purification of phosphoketolase activity.
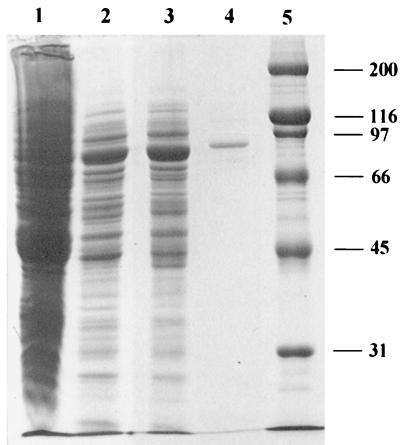
To approach the chemical analysis of the N-terminal amino acid sequence, the main F6P cleaving enzyme of B. lactis was purified to homogeneity from the initial crude extract as summarized in Table 2. The protein was purified to a specific activity of 4.28 U/mg of protein. It appeared as a single band after PAGE under nondenaturing conditions (data not shown). The protein band, when cut out from such a polyacrylamide gel, showed F6PPK activity which proved that we had purified a phosphoketolase. Its apparent molecular weight was 550,000 when estimated by gel filtration chromatography on a calibrated Superdex 200 HR column. On SDS-PAGE, in the presence of β-mercaptoethanol, the purified enzyme displayed a single protein band with an apparent molecular mass of approximately 90,000 Da. A densitometric analysis of the stained protein (Fig. 2, lane 4) yielded more than 99% of the stain in the 90,000-Da band. When the protein which had been obtained from the single band under nondenaturing PAGE was subjected to SDS-PAGE, it migrated as a single band at the 90,000-Da position. These observations suggest that the native enzyme is a homohexamer (6 × 90,000 = 540,000).
FIG. 2.
SDS-PAGE of phosphoketolase preparations from B. lactis. Lanes: 1, crude extract; 2, the sample after DEAE-chromatography; 3, the sample after the Mono Q column; 4, the enzyme after the Superdex 200 column; 5, protein standard. Numbers indicate the molecular mass (in kilodaltons) of the standard proteins. The gel (10% polyacrylamide) was Coomassie stained.
Dual substrate specificity.
To find out whether we had purified the F6P specific phosphoketolase or the X5P/F6P enzyme reported in animal bifidobacteria (31), the apparent Michaelis constants (Km values) for both substrates were analyzed separately. A Michaelis-Menten saturation kinetic was observed for F6P, whereas the kinetics for X5P presented a substrate inhibition profile (7) at X5P concentrations above 20 mM. From double-reciprocal plots apparent Km values of 10 ± 1 mM and 45 ± 3 mM were obtained for F6P and X5P, respectively. Therefore, the purified protein belongs to the class of nonspecific phosphoketolases combining substrate specificity of both phosphoketolase-1 and phosphoketolase-2 (2). The abbreviation Xfp was chosen. The activity of the purified enzyme was not significantly changed by addition of 1 mM ThDP or 2 mM MgCl2 (or both together) to the assay mixture, nor was it changed by omitting sodium fluoride or sodium iodoacetate from the assay. From the initial slopes of the reaction velocities, Vmax values of 5.2 and 27 μmol per min per mg protein were estimated for F6P and X5P, respectively.
N-terminal amino acid sequence of the purified phosphoketolase.
The N-terminal amino acid sequence of the purified enzyme was determined in parallel for the protein after both SDS-PAGE and PAGE under nondenaturing conditions. The first 30 residues were recorded by automatic Edman degradation and were found to be identical in both sources: NH2-XXNPVIGTPWQKLDRPVVEEAIIGMDKYXRV (see also Fig. 1A). This finding supports the homohexamer structure of the native enzyme (see above).
Identification and cloning of the phosphoketolase gene (xfp).
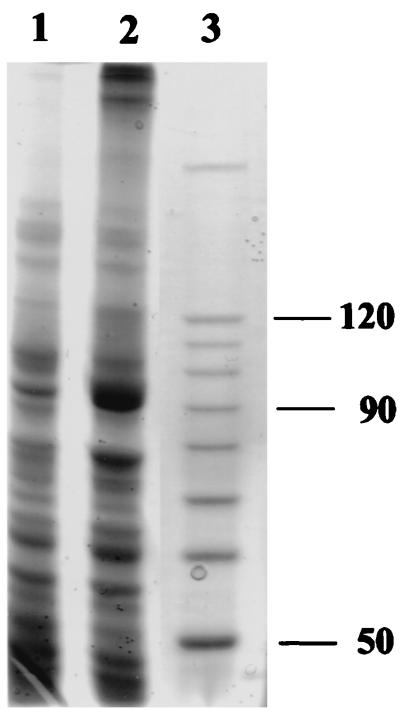
The N-terminal amino acid sequence obtained was used for the identification and cloning of the xfp gene. We applied touchdown PCR with two synthetic oligonucleotide primers (pk5 and pk6). They contain several degeneracies and were designed to target a gene segment for 22 amino acids (Fig. 1). In addition, pk6 has a tail of 9 nucleotides (nt) to prolong the desired PCR product to a length of 75 (Fig. 1). Its DNA sequence confirmed that part of the N terminus encoding DNA had been generated. A Southern blot of B. lactis chromosomal DNA cleaved with BamHI and hybridized with 32P-labeled pk7 (Fig. 1; Table 1) gave a signal of 2.6 kb (data not shown). The proper BamHI fragment and an adjacent BamHI-SmaI fragment (Fig. 1) were finally cloned as a contiguous 3.4-kb fragment in pFPK4 (Table 1) as outlined in Materials and Methods. For this purpose, the low-copy-number plasmid pCL1920 (Table 1) was used as attempts to clone the fragment into the high-copy-number plasmid pUC18 failed. However, no F6PPK activity could be detected in extracts of E. coli containing pFPK4. Therefore, the xfp gene (nt 956 to 3433) was cloned into the expression vector pET-28a(+) by a PCR approach to construct pFPK5 (Table 1). But no F6PPK activity could be measured in extracts of IPTG-induced cultures of the E. coli host BL21(DE3)/pLysS, not even when 1 mM ThDP and 2 mM MgCl2 were added. However, inspection of extracts from such cells revealed a band at a mass of approximately 90,000 Da after SDS-PAGE (see Fig. 5) and indicated the overproduction of the Xfp subunit at the expected size.
FIG. 5.
Expression of the xfp gene from B. lactis in E. coli. Protein extracts of uninduced (lane 1) and IPTG-induced (lane 2) cells from E. coli BL21(DE3)/pLysS harboring plasmid pFPK5 were subjected to SDS-PAGE. The masses (in kilodaltons) of the corresponding protein standards run in lane 3 are indicated at right. The gel (7.5 % polyacrylamide) was Coomassie stained.
DNA sequence of the xfp gene and its adjacent region.
The nucleotide sequence for a 4.1-kb segment of chromosomal B. lactis DNA was derived from sequencing the insertions of both plasmids pFPK1 and pFPK2. In addition, the original arrangement of both insertions on the chromosome was verified by sequence analysis of an overlapping PCR fragment. It had been generated in step 2 of the cloning protocol using pk9 as one of the PCR primers (Fig. 1). The nucleotide sequence of 4,123 bp contained an open reading frame (nt 956 to 3,430) of 825 amino acids. Its deduced N-terminal sequence matches that of the N terminus of the purified phosphoketolase (except for serine-18 [Fig. 1]). The molecular mass of the acidic Xfp polypeptide (pI 4.9) was calculated to be 92,529 Da, a value coincident with that estimated by comparative SDS-PAGE (about 90,000 Da [Fig. 2]). The G+C content of the DNA (61.9%) corresponds exactly to that of the entire B. lactis genome (22). A putative ribosome binding site (AGGAGC) is present 10 to 5 nt upstream of the translational start of the Xfp polypeptide. Hybridization analysis indicated that one single copy of xfp is present on the B. lactis chromosome.
The xfp gene is flanked by two truncated open reading frames (Fig. 1). After aligning the translated regions with those from other microorganisms, the corresponding truncated genes were named guaA and pta, standing for guanosine monophosphate synthetase (EC 6.3.5.2) and phosphotransacetylase (EC 2.3.1.8), respectively. The predicted partial guaA and pta gene products show a 61% identity with the hypothetical guanosine monophosphate synthetase and a 32.2% identity with the phosphotransacetylase reported in the Mycobacterium tuberculosis genome (accession no. Q50729).
Transcriptional analysis of the xfp gene.
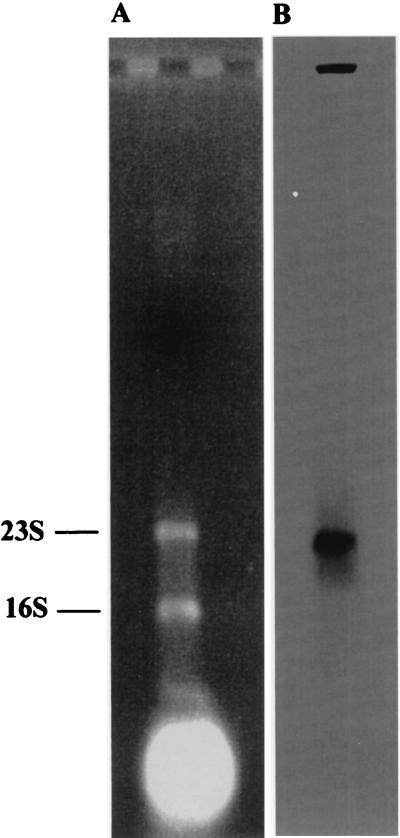
The probe for Northern blot analysis consisted of an internal fragment of the xfp gene (nt 1,385 to 2,119). Hybridizations of total RNA extracted from exponentially growing B. lactis cells with this probe revealed a single distinct signal indicating an mRNA transcript of approximately 2,500 nt (Fig. 3). Thus, the xfp gene is not cotranscribed with other adjacent genes. Two perfect inverted repeats (each 13 nt in length) were detected 25 nt downstream of the translational end of xfp (nt 3,459 to 3,471 and 3,476 to 3,488, respectively). They could build up transcriptional terminator structures in B. lactis. Potential −35 (AGGTCA, nt 729 to 734) and −10 (CATAAT, nt 849 to 854) promoter motifs are located upstream of a GA dinucleotide (nt 859 to 860). Either of these particular G or A positions represents a transcriptional start point of the xfp gene (starting at position 956) as we found in preliminary primer extension experiments (data not shown).
FIG. 3.
Northern blot analysis of B. lactis xfp gene transcript. Ethidium bromide staining of RNA from B. lactis after agarose gel electrophoresis (A) and X-ray signal after Northern blot analysis using primer pk9 as a probe (B). 16S and 23S indicate the migrating positions of 16S and 23S rRNA, respectively.
DISCUSSION
The purified phosphoketolase Xfp from B. lactis has Km values of 10 mM (for F6P) and 45 mM (for X5P). The calculated Vmax values for F6P and X5P are 5.2 and 27 μmol per min per mg of protein, yielding a ratio of 1:5. Xfp of B. lactis is similar to a phosphoketolase partially purified from Bifidobacterium globosum, whose habitat is the bovine rumen (31). The typical feature of the B. globosum enzyme was reported to be a high reaction rate with X5P compared to F6P. In this respect, the enzyme resembles also the phosphoketolase from Acetobacter xylinum (30). Also, the crystallized phosphoketolase from Leuconostoc mesenteroides accepts both substrates with Km values of 4.7 mM (X5P) and 29 mM (F6P), respectively (10).
The molecular weight of the native enzyme from B. lactis is 550,000, compared to 290,000 (31) or 300,000 (12) estimated by gel filtration for the B. globosum enzyme. The X5P specific enzyme from Lactobacillus plantarum had a molecular weight of 550,000 as determined by ultracentrifugation and the resulting Svedberg constant (13). Our homohexamer hypothesis for the B. lactis enzyme was derived from the genetically or electrophoretically determined subunit size of 92,529 Da and the calibrated gel filtration experiment. It will have to be substantiated by additional biochemical evidence such as controlled chemical cross-linking followed by chromatographic molecular weight estimations. Furthermore, the size and subunit composition of the Xfp protein is totally different from the αβ subunit structure of a recently described F6PPK activity (molecular weight, 110,000 to 115,000) purified from B. asteroides extracts (K. G. Fandi, H. M. Ghazali, and A. M. Yazid, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., p. 436, 2000). Our data do not allow yet conclusions as to whether the enzyme is involved in both steps (phosphoketolase-1 and -2) of the so-called Bifidobacterium bifidum pathway (2). It should be mentioned that the crude extract of B. lactis contains a second phosphoketolase activity which can be separated by gel filtration from Xfp. It seems to be specific for F6P (L. Rohr, unpublished observation).
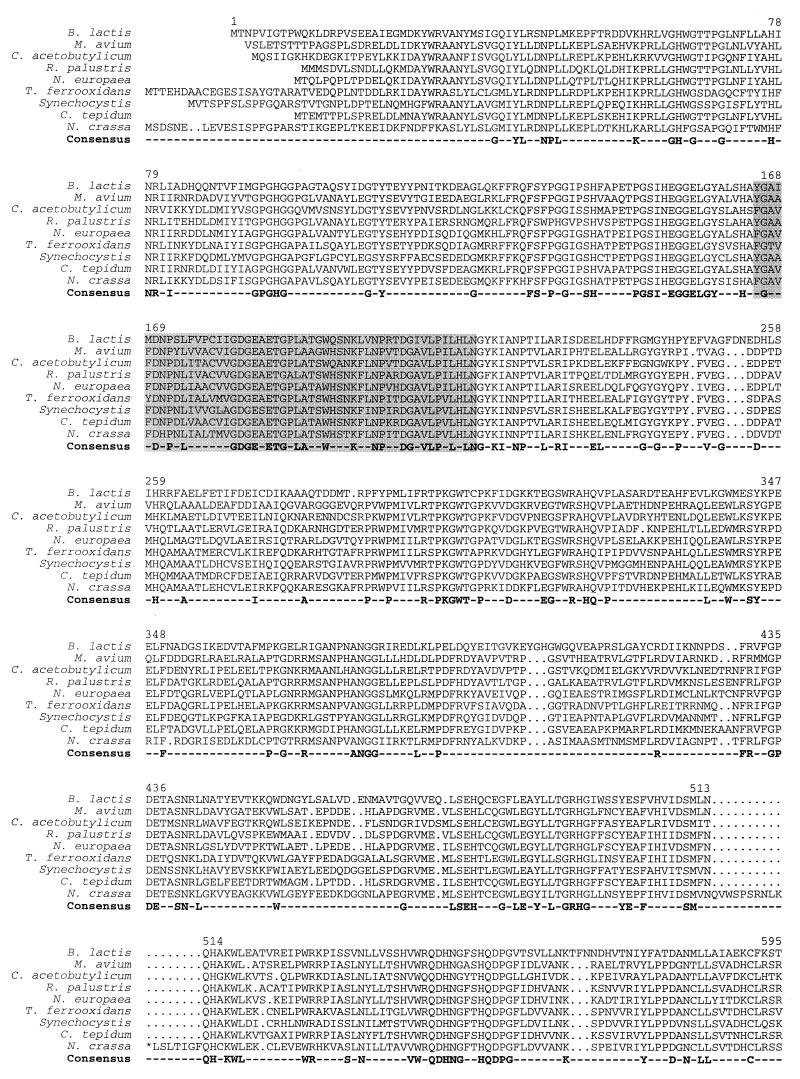
Although the phosphoketolase pathway has been proposed in the past to be specific for a small group of bacteria and yeasts, sequences highly homologous to xfp of B. lactis were detected in the genomes from a variety of eubacteria and eucarya, but not from archaea. On the protein level, the calculated amino acid sequence identities vary between 26 and 55%; similarities range up to 63%. The taxonomic diversity is represented in the alignment shown in Fig. 4.
FIG. 4.
Alignment of the B. lactis Xfp amino acid sequence with homologous hypothetical proteins identified in the chromosomes of Chlorobium tepidum (The Institute for Genomic Research [TIGR]), C. acetobutylicum (Genome Therapeutics), Mycobacterium avium (TIGR), Neurospora crassa (Heinrich Heine Universität Düsseldorf), Nitrosomonas europaea (Doe Joint Genome Institute), Rhodopseudomonas palustris (Doe Joint Genome Institute), Synechocystis sp. strain PCC6803 (P74690), and Thiobacillus ferrooxidans (TIGR) (sequence sources are given in parenthesis). Amino acid residues that occur in all of the 9 sequences are included in a consensus sequence. The thiamine diphosphate binding motif is shadowed. Amino acid numbering refers to the B. lactis Xfp protein. Further homologies were found in the chromosomes of Anabaena sp. strain PCC7120 (two different copies) (Kazusa DNA Research Institute), Brucella suis (TIGR), Lactococcus lactis IL1403 (Genoscope), Mesorhizobium loti (AP003009), Mycobacterium smegmatis (TIGR), Nostoc punctiforme (Doe Joint Genome Institute), Pseudomonas aeruginosa (Pseudomonas Genome Project), Pseudomonas syringae pv. tomato (TIGR), Schizosaccharomyces pombe (SPBC24C6), Shewanella putrefaciens (TIGR), Sinorhizobium meliloti (Stanford University), and Streptomyces coelicolor (SCF55/SCF56).
In Clostridium acetobutylicum ATCC 824, the xfp-homologous gene (coding for open reading frame CAC1622) is situated in a cluster of genes for the pentose phosphate pathway and some anaplerotic reactions: araE, araR, l-ribulose phosphate 4-epimerase, l-arabinose isomerase, xfp, xylulose kinase, ywtG (metabolite transport), l-arabinose isomerase, transaldolase, transketolase, and an aldose 1-epimerase (nt 1,477,589 to 1,494,444 of the chromosome) (data from Genomic Therapeutics Corp.).
The amino acid sequence contains a ThDP-dependent enzyme signature sequence Y-G-X5-P-X3-V-X2-I-X-G-D-G-E (amino acids 165 to 184 in Xfp) matching closely the consensus pattern [LIVMF]-G-X5-P-X4-V-X-I-X-G-D-G-[GSAC] (PROSITE PS00187 [Fig. 4]). Another motif, G-D-G-X24–27-N-N, of ThDP-binding enzymes like acethydroxyacid synthases, transketolases, E1 components of 2-ketoacid and acetoin dehydrogenases, and others (4) is also present in Xfp in a slightly modified form (G-D-G-E-X30-N) (amino acids 181 to 215 [Fig. 4]).
In the course of cloning the B. lactis xfp gene in E. coli, we failed to clone the entire gene in the high-copy-number vector pUC18. It might point to the effect that the basic carbohydrate pathways of E. coli might be severely disturbed by xfp and its expression, leading to a lethal imbalance of metabolites (34). We failed to detect F6PPK activity in E. coli harboring the recombinant xfp gene in both the low-copy-number vector pCL1920 and in the expression vector pET-28a(+), respectively (Table 1). However, the Xfp subunit seemed to be synthesized in the E. coli host carrying pFPK5 (Fig. 5). Due to the additional His tag arrangement (36 amino acids encoded by the expression vector) at the N terminus of the recombinant Xfp, the functionality was probably hampered by an inappropriate subunit assembling to the hexamer. Although expression of an active enzyme in E. coli is still pending, the biochemical evidence that the characterized gene xfp codes for the purified Xfp is sufficiently substantiated.
ACKNOWLEDGMENTS
We thank Alexandra Kulangara for help during the purification of the protein.
This work was supported by grant 01016/41-2705.5 from ETH Zurich.
REFERENCES
- 1.Biavati B, Sgorbati B, Scardovi V. The genus Bifidobacterium. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1991. pp. 816–833. [Google Scholar]
- 2.Buckel W. Anaerobic energy metabolism. In: Lengeler J W, Drews G, Schlegel H G, editors. Biology of the prokaryotes. Stuttgart, Germany: Thieme-Verlag; 1999. pp. 278–326. [Google Scholar]
- 3.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactoside selection. BioTechniques. 1987;4:376–379. [Google Scholar]
- 4.Candy J M, Duggleby R G. Structure and properties of pyruvate decarboxylase and site-directed mutagenesis of the Zymomonas mobilis enzyme. Biochim Biophys Acta. 1998;1385:323–338. doi: 10.1016/s0167-4838(98)00077-6. [DOI] [PubMed] [Google Scholar]
- 5.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis B J. Disc gel electrophoresis-II. Method and application to human serum proteins. Ann N Y Acad Sci. 1964;121:404–424. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 7.Dixon M, Webb E C. Enzymes. 2nd ed. London, United Kingdom: Longmans; 1964. p. 81. [Google Scholar]
- 8.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldberg M L, Racker E. Formation and isolation of a glycolaldehyde-phosphoketolase intermediate. J Biol Chem. 1962;237:3841–3842. [PubMed] [Google Scholar]
- 10.Goldberg M, Fessenden J M, Racker E. Phosphoketolase. Methods Enzymol. 1966;9:515–520. [Google Scholar]
- 11.Greenley D E, Smith D W. A novel pathway of glucose catabolism in Thiobacillus novellus. Arch Microbiol. 1979;122:257–262. [Google Scholar]
- 12.Grill J-P, Crociani J, Ballongue J. Characterization of fructose 6-phosphate phosphoketolases purified from Bifidobacterium species. Curr Microbiol. 1995;31:49–54. doi: 10.1007/BF00294634. [DOI] [PubMed] [Google Scholar]
- 13.Heath E C, Hurwitz J, Horecker B L, Ginsburg A. Pentose fermentation by Lactobacillus plantarum. I. The cleavage of xylulose 5-phosphate by a phosphoketolase. J Biol Chem. 1958;231:1009–1029. [PubMed] [Google Scholar]
- 14.Holzer H, Schröter W. Zum Wirkunsmechanismus der Phosphoketolase. I. Oxydation verschiedener Substrate mit Ferricyanid zu Glycolsäure. Biochim Biophys Acta. 1962;65:271–288. doi: 10.1016/0006-3002(62)91046-6. [DOI] [PubMed] [Google Scholar]
- 15.Kandler O. Carbohydrate metabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1983;49:209–224. doi: 10.1007/BF00399499. [DOI] [PubMed] [Google Scholar]
- 16.Kullen M J, Klaenhammer T R. Genetic modification of intestinal lactobacilli and bifidobacteria. In: Tannock G W, editor. Probiotics—a critical review. Wymondham, United Kingdom: Horizon Scientific Press; 1999. pp. 65–83. [Google Scholar]
- 17.Leenhouts K J, Kok J, Venema G. Campbell-like integration of heterologous plasmid DNA into the chromosome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989;55:394–400. doi: 10.1128/aem.55.2.394-400.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmann Y, Meile L, Teuber M. Rubrerythrin from Clostridium perfringens: cloning of the gene, purification of the protein, and characterization of its superoxide dismutase function. J Bacteriol. 1996;178:7152–7158. doi: 10.1128/jb.178.24.7152-7158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 21.Meile L, Stettler R, Banholzer R, Kotik M, Leisinger T. Tryptophan gene cluster of Methanobacterium thermoautotrophicum Marburg: molecular cloning and nucleotide sequence of a putative trpEGCFBAD operon. J Bacteriol. 1991;173:5017–5023. doi: 10.1128/jb.173.16.5017-5023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meile L, Ludwig W, Rueger U, Gut C, Kaufmann P, Dasen G, Wenger S, Teuber M. Bifidobacterium lactis sp. nov., a moderately oxygen tolerant species isolated from fermented milk. Syst Appl Microbiol. 1997;20:57–64. [Google Scholar]
- 23.Oelmüller U, Krüger N, Steinbüchel A, Friedrich C G. Isolation of procaryotic RNA and detection of specific mRNA with biotinylated probes. J Microbiol Methods. 1990;11:73–84. [Google Scholar]
- 24.Orban J I, Patterson J A. Modification of the phosphoketolase assay for rapid identification of bifidobacteria. J Microbiol Methods. 2000;40:221–224. doi: 10.1016/s0167-7012(00)00133-0. [DOI] [PubMed] [Google Scholar]
- 25.Pearson P W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Racker E. Fructose-6-phosphate posphoketolase from Acetobacter xylinum. Methods Enzymol. 1962;5:276–280. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schörken U, Sprenger G A. Thiamin-dependent enzymes as catalysts in chemoenzymatic syntheses. Biochim Biophys Acta. 1998;1385:229–243. doi: 10.1016/s0167-4838(98)00071-5. [DOI] [PubMed] [Google Scholar]
- 30.Schramm M, Klybas V, Racker E. Phosphorolytic cleavage of fructose-6-phosphate by fructose-6-phosphate phosphoketolase from Acetobacter xylinum. J Biol Chem. 1958;233:1283–1288. [PubMed] [Google Scholar]
- 31.Sgorbati B, Lenaz G, Casalicchio F. Purification and properties of two fructose 6-phosphate phosphoketolases in Bifidobacterium. Antonie Leeuwenhoek. 1976;42:49–57. doi: 10.1007/BF00399448. [DOI] [PubMed] [Google Scholar]
- 32.Veiga-da-Cunha M, Santos H, van Schaftingen E. Pathway and regulation of erythritol formation in Leuconostoc oenos. J Bacteriol. 1993;175:3941–3948. doi: 10.1128/jb.175.13.3941-3948.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate polyacrylamide gel electrophoresis. J Biol Chem. 1961;244:4406–4412. [PubMed] [Google Scholar]
- 34.Wei B, Shin S, Laporte D, Wolfe A J, Romeo T. Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. J Bacteriol. 2000;182:1632–1640. doi: 10.1128/jb.182.6.1632-1640.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC18 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]