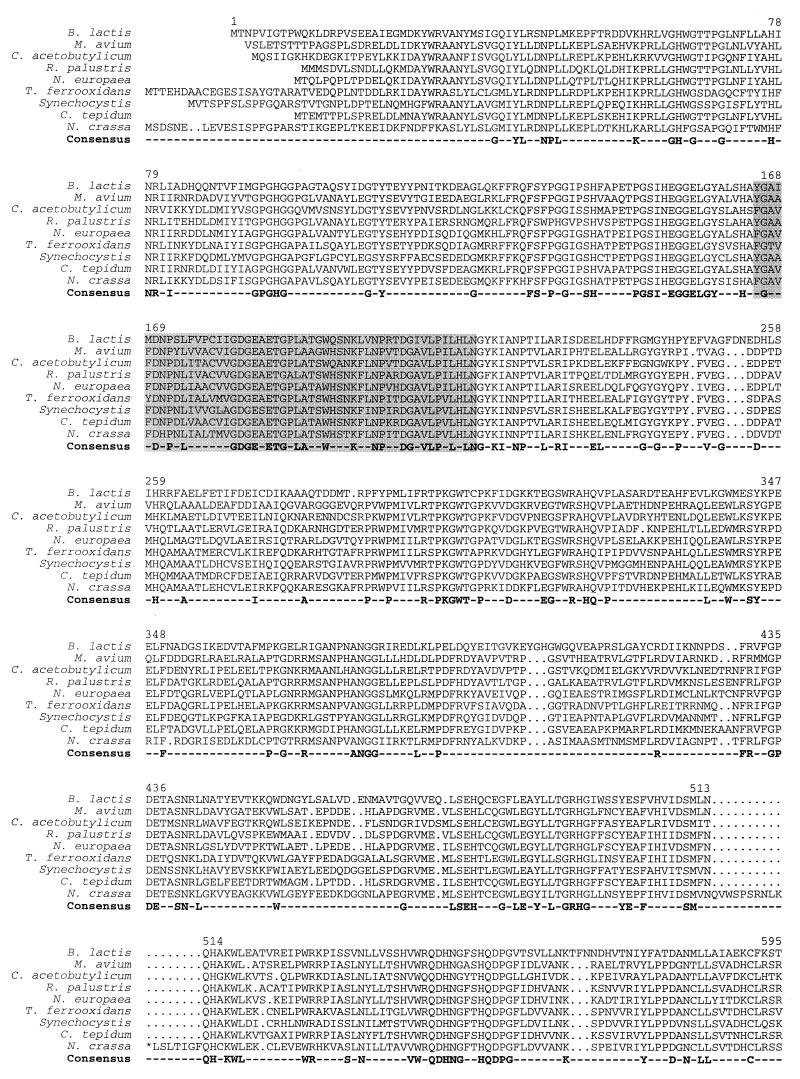
FIG. 4.
Alignment of the B. lactis Xfp amino acid sequence with homologous hypothetical proteins identified in the chromosomes of Chlorobium tepidum (The Institute for Genomic Research [TIGR]), C. acetobutylicum (Genome Therapeutics), Mycobacterium avium (TIGR), Neurospora crassa (Heinrich Heine Universität Düsseldorf), Nitrosomonas europaea (Doe Joint Genome Institute), Rhodopseudomonas palustris (Doe Joint Genome Institute), Synechocystis sp. strain PCC6803 (P74690), and Thiobacillus ferrooxidans (TIGR) (sequence sources are given in parenthesis). Amino acid residues that occur in all of the 9 sequences are included in a consensus sequence. The thiamine diphosphate binding motif is shadowed. Amino acid numbering refers to the B. lactis Xfp protein. Further homologies were found in the chromosomes of Anabaena sp. strain PCC7120 (two different copies) (Kazusa DNA Research Institute), Brucella suis (TIGR), Lactococcus lactis IL1403 (Genoscope), Mesorhizobium loti (AP003009), Mycobacterium smegmatis (TIGR), Nostoc punctiforme (Doe Joint Genome Institute), Pseudomonas aeruginosa (Pseudomonas Genome Project), Pseudomonas syringae pv. tomato (TIGR), Schizosaccharomyces pombe (SPBC24C6), Shewanella putrefaciens (TIGR), Sinorhizobium meliloti (Stanford University), and Streptomyces coelicolor (SCF55/SCF56).