Abstract
Since the beginning of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)/coronavirus disease 2019 (COVID-19) pandemic, global sequencing efforts have led in the field of inborn errors of immunity, and inspired particularly by previous research on life-threatening influenza, they have revealed that known and novel inborn errors affecting type I interferon immunity underlie critical COVID-19 in up to 5% of cases. In addition, neutralizing autoantibodies against type I interferons have been identified in up to 20% of patients with critical COVID-19 who are older than 80 years and 20% of fatal cases, with a higher prevalence in men and individuals older than 70 years. Also, inborn errors impairing regulation of type I interferon responses and RNA degradation have been found as causes of multisystem inflammatory syndrome in children, a life-threatening hyperinflammatory condition complicating otherwise mild initial SARS-CoV-2 infection in children and young adults. Better understanding of these immunologic mechanisms can aid in designing treatments for severe COVID-19, multisystem inflammatory syndrome in children, long COVID, and neuro-COVID.
Key words: COVID-19, SARS-CoV-2, multisystem inflammatory syndrome in children, type I interferon
In December 2019, the coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged as a novel human pathogen causing coronavirus 2019 disease (COVID-19), an infection characterized by pneumonia and acute respiratory failure in a significant proportion of cases (5%-15%).1, 2, 3 Like other respiratory viruses, SARS-CoV-2 enters and infects respiratory epithelial cells, mostly by binding to the angiotensin-converting enzyme 2 (ACE2) receptor, where it replicates. It can then spread to other organs, mainly through a viremic phase. The detection of viral components through pattern recognition receptors (including Toll-like receptors [TLRs], retinoic acid–inducible gene I [RIG-I]-like receptors, and nucleotide-binding oligomerization domain [NOD]-like receptors [NLRs]) and cytosolic sensors (such as cyclic guanosine monophosphate–AMP synthase), activates an antiviral response in infected epithelial cells as well as in leukocytes governing the innate immune response, such as macrophages, monocytes, dendritic cells, neutrophils, and innate lymphoid cells.4 , 5 Type I and type III interferon signal through the interferon receptors (IFNAR1/2 for type I interferon and IFNLR1/IL10RB for type III interferon) and signal transducer and activator of transcription (STAT) 1 and 2, which combine with interferon regulatory factor 9 (IRF9) to induce the expression of interferon-stimulated genes responsible for antiviral defense. The interferon response needs to be finely tuned to strike a balance between virus clearance and prevention of excessive inflammation, which can be further exacerbated by the viral-induced activation of the inflammasome and cause a cytokine storm mediated by inflammatory cell death.5
From the start of the pandemic, around 0.5% to 1% of patients died and 2% to 4% experienced critical disease globally.6 , 7 Risk factors for critical COVID-19 are age (with a doubling of the risk every 5 years of age), male sex, and comorbidities such as obesity, type 2 diabetes, and chronic lung disease.7 , 8 However, the impact of the comorbidities in terms of odds ratio is at best limited and does not explain the striking interindividual variability in severity of disease following SARS-CoV-2 infection. In an attempt to explain this variability, several international consortia have searched for rare or common human genetic variants on a large scale that could modify the risk of infection or of severe COVID-19.9, 10, 11, 12, 13, 14 In particular the COVID Human Genetic Effort (www.COVIDHGE.com) has made landmark discoveries to explain critical COVID-19. Consequently, a better understanding of the key players in human defense against SARS-CoV-2 led to the finding of other, acquired, immunologic risk factors. Finally, research has also concentrated on puzzling complications of COVID-19, such as multisystem inflammatory disease in children (MIS-C), long COVID, and “COVID toes.” Here, we review the findings that have cast light on defects of innate immunity causing susceptibility to COVID-19 and MIS-C (summarized in Table I, Table II, Table III 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36 and Fig 1). The implication of these findings on the implementation of targeted treatment in patients with inborn errors of immunity or their phenocopies is beyond the scope of this review, and we refer readers to a previous review on this topic. 37
Table I.
Inborn errors of type I interferon immunity underlying susceptibility to severe COVID pneumonia
| Gene | Inheritance | Other known phenotypes | Reference(s) |
|---|---|---|---|
| IRF7 | AR | Influenza pneumonia | 15, 16, 17 |
| AD | NA | ||
| IFNAR1 | AR | Fatal susceptibility to live viral vaccines, HLH | 15,18, 19, 20 |
| AD | NA | ||
| TLR7 | XLR | NA | 20, 21, 22, 23 |
| TLR3 | AD | Influenza pneumonia, herpes simplex encephalitis | 15,24 |
| UNC93B1 | AD | NA (AR herpes simplex encephalitis) | 15 |
| TICAM1 | AD | Herpes simplex encephalitis | 15 |
| TBK1 | AD | Herpes simplex encephalitis | 15,25 |
| IRF3 | AD | Herpes simplex encephalitis | 15 |
| IFNAR2 | AD | NA (AR fatal susceptibility to live viral vaccines) | 15 |
| STAT2 | AR | Influenza pneumonia, fatal susceptibility to live viral vaccines, HLH | 20 |
| TYK2 | AR | Susceptibility to mycobacteria and viruses, hyper-IgE syndrome | 20 |
| MYD88 and IRAK4 | AR | Pyogenic infections | 26, 47 |
Mechanism is disruption of an essential mediator of type I interferon responses. Proportion of severe COVID pneumonia in young patients (<60 years) explained in 1% to 5% of cases.
AD, Autosomal dominant; AR, autosomal recessive; NA, not applicable; XLR, X-linked recessive.
Table II.
Autoimmune phenocopies of type I interferon defects underlying susceptibility to severe COVID pneumonia
| Phenocopies of type I interferon defects | Reference(s) |
|---|---|
| Patients treated with recombinant IFN-α or IFN-β Patients with autoimmune conditions:
|
27, 28, 29, 30, 31, 32, 33 |
Mechanism is neutralizing autoantibodies against type I IFN. Proportion of critical COVID pneumonia is explained in 15% to 20% of cases.
IPEX, Immunodysregulation polyendocrinopathy enteropathy X-linked; SLE, systemic lupus erythematosus; XL, X-linked.
Table III.
Inborn errors of immunity underlying susceptibility to MIS-C
| Gene | Inheritance | Other known phenotypes | Reference(s) |
|---|---|---|---|
| XIAP | XL | Familial HLH | 34 |
| CYBB | XL | Chronic granulomatous disease | 14,34 |
| SOCS1 | AD | Early-onset familial autoimmunity | 34,35 |
| OAS1 | AR | NA | 36 |
| OAS2 | AR | NA | 36 |
| RNASEL | AR | NA | 36 |
Mechanism is excessive inflammatory responses to SARS-CoV-2 due to defective viral RNA degradation and/or dysregulated interferon and inflammasome activation.
AD, Autosomal dominant; AR, autosomal recessive; NA, not applicable; XL, X-linked.
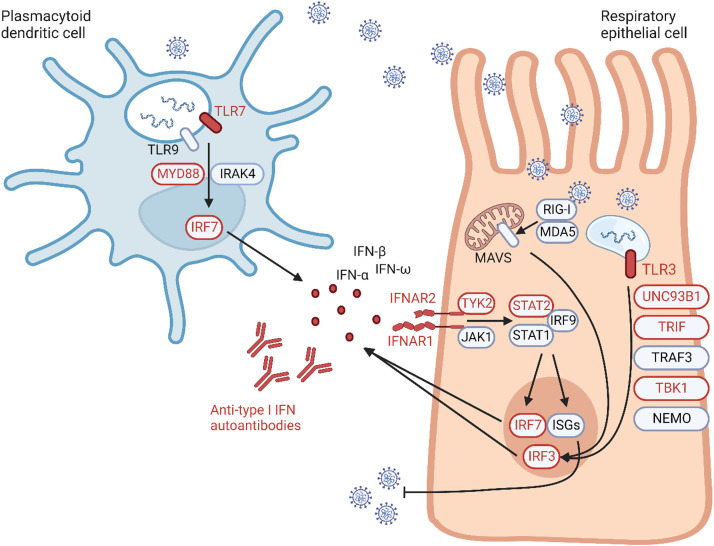
Fig 1.
Inborn errors of type I interferon (IFN) immunity and their phenocopies causing susceptibility to severe COVID-19. The activation of the type I IFN responses in respiratory epithelial cells and plasmacytoid dendritic cells following infection with SARS-CoV-2 is illustrated. Viral particles processed through endosomes activate TLR3, TLR7, and TLR9. TLR3 signals through UNC93B1, TRIF, TRAF3, TBK1, and NEMO to induce IRF3. IRF3 is also activated by RIG-I and MDA5, which signal through mitochondrial antiviral signaling (MAVS) following intracellular sensing of viral nucleic acids. TLR7 and TLR9 signal instead via MYD88 and IRAK4 to induce IRF7. Both IRF7 and IRF3 are transcription factors driving the production of type I IFN that in turn bind to their receptor and signal through the STAT1-STAT2-IRF9 complex to induce IRF7 and the transcription of interferon-stimulated genes, which have broad antiviral activities. Monogenic inborn errors of genes involved in these responses were found in patients with critical or fatal COVID-19 and are indicated in red. A phenocopy of these inborn errors is represented by neutralizing autoantibodies against type I IFN, which is also found in a significant proportion of patients with severe COVID-19. (Created with BioRender.)
Monogenic type I interferon defects and severe COVID-19
The initial focus of the COVID Human Genetic Effort was centered on inborn errors of type I interferon immunity in patients with life-threatening COVID-19.15 , 27 , 38 , 39 SARS-CoV-2 and influenza are both RNA viruses affecting the respiratory tract and causing life-threatening pneumonia. Thus, it was hypothesized that predisposition to critical COVID-19 and influenza could be allelic. Zhang et al therefore analyzed 3 genetic loci underlying influenza susceptibility (TLR3, IRF7, and IRF9) and 10 additional closely related loci involved in antiviral responses (TRIF, TRAF3, TBK1, UNC93B1, IRF3, STAT1, STAT2, IFNAR1, IFNAR2, and NEMO) in 659 patients of all ages with critical COVID-19 and a control group of 534 individuals with mild or asymptomatic COVID-19.15 They found an enrichment in monoallelic and biallelic pathogenic variants in 8 genes (TLR3, UNC93B1, TRIF, TBK1, IRF3, IRF7, IFNAR1, and IFNAR2) in the patients but not in the control group. A defect of TLR3- and IRF7-dependent type I interferon immunity was demonstrated in cells with complete IRF7 and IFNAR1 deficiency, as were an impaired intracellular response to infection with SARS-CoV-2 in vitro and significantly lower serum type I interferon levels in vivo.15 These findings were confirmed by several additional reports of autosomal recessive IRF7, IFNAR1, TBK1, and TYK2 deficiency in patients with critical COVID pneumonia, including in children.16 , 18, 19, 20 , 25
Subsequently, the hypothesis that an enrichment of deleterious variants in the X chromosome could explain the higher proportion of males affected by severe COVID-19 was tested by using an unbiased approach.21 This led to the discovery of X-linked recessive TLR7 deficiency in about 2% of males with critical COVID-19 pneumonia, including children.20, 21, 22, 23 The penetrance of severe SARS-CoV-2 infection appears to be high, but not complete, for the described recessive defects and lower for dominant defects. Interestingly, patients with autosomal recessive IFNAR1 or IRF7 deficiency had not previously developed severe disease following live viral vaccine administration, influenza virus infection, or other viral infections,40, 41, 42, 43, 44, 45 as has been described for earlier cases of individuals with pathogenic biallelic variants in these genes. This observation illustrates the variable expression of the phenotype as well as the redundancy of type I interferon responses for human defense against most viruses. Finally, some cases of critical SARS-CoV-2 pneumonia in patients with myeloid differentiation factor 88 (MyD88) or IRAK4 deficiency have also been reported.26 , 46 , 47 MyD88 and IRAK4 are essential mediators of signaling downstream from TLR7, and a role for MyD88 in controlling pulmonary replication of SARS-CoV-1 in mice was previously shown.48 Also, impaired type I/III interferon production following SARS-CoV-2 infection was shown in vitro in plasmacytoid dendritic cells from a patient with IRAK4 deficiency.49
The relevance of interferon-mediated immunity in the context of COVID-19 has also been confirmed by the findings of several population-based genome-wide association studies. For example, a recent genome-wide gene-based rare variant association analysis confirmed enrichment in rare loss-of-function variants in TLR7, TYK2, and several IRF7- and TLR3-dependent type I interferon immunity loci50; in addition, other genome wide association studies have demonstrated significant association of critical infection with variants involved in interferon signaling (IFNAR2, TYK2, CCR2, and IL10RB) and cytosolic double-stranded RNA (dsRNA) sensing (2’-5’-oligoadenylate synthetase 1 gene [OAS1], OAS2, and OAS3).11 , 51 Other significant effects were found for loci related to the blood group and within the 3p21.31 region (possibly affecting ACE2 expression) regarding susceptibility to infection with SARS-CoV-2, and several loci related to lung disease regarding risk of severe infection.10 , 11
Autoimmune phenocopies of type I interferon defects
Monogenic defects of antiviral immunity explain only a minor proportion of the patients with critical COVID-19 in the population younger than 60 years (estimated as 1%-5%).52 The hypothesis that the presence of serum neutralizing autoantibodies against type I interferons could mimic these inborn errors in a larger proportion of severely affected patients was then raised. This phenocopy mechanism is well known in the field of immunodeficiencies. Indeed, autoimmune phenocopies of mendelian susceptibility to mycobacterial disease, invasive pneumococcal disease, and chronic mucocutaneous candidiasis have been attributed to neutralizing autoantibodies against the cytokines IFN-γ, IL-6, and IL-17A/F, respectively.53, 54, 55, 56, 57 The presence of autoantibodies against type I interferon in patients with a variety of conditions, inborn and acquired, has been known for several decades. These include patients with elevated levels of type I interferon, such as those receiving therapy with IFN-α or IFN-β for multiple sclerosis or patients with SLE, patients with broad autoimmune diseases (such as thymoma and myasthenia gravis), and patients with immune defects of central or peripheral tolerance (such as autoimmune polyendocrinopathy syndrome 1, combined immunodeficiency with autoimmunity due to mutations in RAG1 or RAG2, and immunodysregulation polyendocrinopathy enteropathy X-linked syndrome).58, 59, 60, 61, 62, 63, 64
The clinical relevance of anti–type I interferon autoantibodies was unclear for several decades, and no association with viral infections was reported, except in a patient with varicella-zoster and some patients with RAG1 or RAG2 deficiency and severe chickenpox.60 , 65 , 66 Indeed, although the presence of anti–type I interferon autoantibodies was being utilized as a diagnostic criterion for autoimmune polyendocrinopathy syndrome 1 (APS-1) and an inverse correlation between high titers of these antibodies and type 1 diabetes had been described, a clinical correlate for increased susceptibility for infection was lacking.67 At the beginning of the pandemic, an early report illustrated the critical course of COVID-19 pneumonia in an Italian patient with APS-1.68 Bastard et al then tested the hypothesis that autoantibodies against type I interferons could underlie severe COVID-19 and initially found that at least 10% of patients of all ages with critical pneumonia had neutralizing autoantibodies against IFN-α, IFN-ω, or both and that the rate of severe or critical pneumonia increased to 86% in patients with APS-1 with these neutralizing antibodies.27 , 28 The study group also confirmed that this resulted in low or undetectable serum IFN-α levels during acute SARS-CoV-2 infection in all patients.27 Strikingly, 94% of these patients were men, which could contribute to the higher prevalence of severe COVID-19 in males.27
The association of neutralizing autoantibodies against type I interferons and critical COVID-19 was confirmed by many independent studies.28 , 29 , 69, 70, 71, 72, 73, 74, 75, 76 It was shown that neutralizing autoantibodies against IFN-α or IFN-ω are rare in the general population (they are present in less than 1% of those between 20 and 70 years old), that their prevalence increases with age (reaching 4% of individuals older than 70 years), and that they are present in about 20% of both patients with critical COVID-19 who are older than 80 years and individuals with fatal cases in all age groups.28 , 29 , 69, 70, 71, 72, 73, 74, 75, 76 The presence of neutralizing anti–type I interferon autoantibodies correlates with an increased risk of death and an increased infection fatality rate in patients infected with SARS-CoV-2, and this risk rises with age.30 This effect is seen both in the general population of patients with COVID-19 and in patients with APS-1, who showed a significantly higher risk of critical and fatal disease than did age-matched patients without the condition.28 Moreover, in a cohort of 48 patients with breakthrough severe COVID-19 pneumonia after 2 doses of an mRNA vaccine, 24% of subjects had autoantibodies neutralizing type I interferons.31 The presence of neutralizing anti–type I interferon autoantibodies could therefore explain the atypically severe infections in vaccinated individuals in at least a quarter of cases.31 A role for anti–type I interferon autoantibodies in other severe viral diseases is being explored further, as these autoantibodies seem to be correlated with severe herpesvirus infections (eg, severe cutaneous herpes zoster, varicella pneumonia, varicella central nervous system vasculitis, and cytomegalovirus infection), life-threatening yellow fever vaccine associated disease, and critical influenza pneumonia in almost 5% of patients.71 , 76, 77, 78, 79, 80 Interestingly, in a recent study of 609 patients with SLE, anti–type I interferon autoantibodies were found in 11.7% of patients regardless of age or sex.71 Only 20 of the 71 samples had neutralizing activity though, and this was significantly associated with episodes of cutaneous herpes zoster and severe viral infection, including increased risk of severe COVID-19. Strikingly, patients with autoantibodies neutralizing several different type I interferons had the highest risk, and these patients were almost uniquely women.71 This is in line with the data presented by Manry et al, which show that the effect of neutralizing autoantibodies against type I interferons on relative risk of death and infection fatality rate are more important than, for instance, maleness.30
MIS-C
Children and young adults are mostly spared from critical COVID-19. However, a few months after the onset of the pandemic, MIS-C emerged as a severe complication usually affecting children a few weeks after a mild or asymptomatic infection with SARS-CoV-2 in regions in which the incidence of SARS-CoV-2 infection was high.81, 82, 83, 84, 85 MIS-C shows a higher prevalence in males and individuals of African or Hispanic ancestry.85 Despite overlapping features with Kawasaki disease, MIS-C is often diagnosed in older children (in children aged 7.5-12 years with MIS-C versus in children younger than 5 years with Kawasaki disease), and it has a more severe course with multiorgan dysfunction in more than 70% of patients and shock and myocarditis in 50% and 90% of patients, respectively.81, 82, 83, 84 The hyperinflammatory state is characterized by elevated levels of cytokines and cytopenia, which often fulfil the diagnostic criteria for hemophagocytic lymphohistiocytosis (HLH).81, 82, 83, 84 As in the case of critical COVID-19 pneumonia, it was hypothesized that specific immune defects could predispose to MIS-C. Early studies reported patients with immune dysregulation syndromes and chronic granulomatous disease (CGD) experiencing extreme inflammation during SARS-CoV-2 infection classified as HLH or MIS-C.14 , 86 Subsequently, a prospective targeted sequencing approach unveiled inborn errors of suppressor of cytokine signaling 1 (SOCS1; n = 2), X-linked inhibitor of apoptosis (XIAP; n = 1) and cytochrome B-245 β-chain (CYBB; n = 1) in a cohort of children with MIS-C.34 , 35 SOCS1 is a negative regulator of type I and II interferon responses that binds Janus kinase 1 and 2, impeding activation of STAT1 and STAT2 downstream of the interferon receptors. SOCS1 haploinsufficiency has been described in subjects with interferon-driven early-onset familial autoimmunity and lymphoproliferation.87 Hemizygous loss-of-function mutations in XIAP cause immune dysregulation characterized by HLH, inflammatory bowel disease, and inflammation.88 Finally, CGD is a neutrophil disorder caused by impairment in 1 of the subunits of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex, which includes the product of CYBB. Patients with CGD are prone to invasive pyogenic and fungal infections and granulomatous inflammation. NADPH oxidase defects also predispose to infection-triggered HLH as well as to other noninfectious autoimmune and autoinflammatory manifestations, probably owing to a substantial influx of neutrophils that, although impaired in the oxidative function, can still trigger a strong inflammatory response.89 , 90 Chou et al showed an increased inflammatory signature in these children, driven primarily by type I and II interferon and nuclear factor-κB–IL-6 responses.34 These preliminary findings point to a predisposition to MIS-C in children with inborn errors of immune dysregulation, especially affecting regulators of interferon responses. The significance of these case studies will need to be validated in larger cohorts of patients.
A whole genome/whole exome sequencing approach was used by the COVID Human Genetic Effort to screen a large cohort of 558 children (aged 0-19 years) with MIS-C. By filtering for rare homozygous or hemizygous variants (allele frequency <0.01) in genes involved in antiviral responses, they identified the OAS-RNase L pathway as a relevant signaling circuit in MIS-C.36 OAS1, OAS2, and OAS3 are interferon-inducible cytosolic dsRNA sensors that activate the endoribonuclease RNase L, which degrades human and viral single-stranded RNA. In this study, 5 patients with autosomal recessive OAS1, OAS2, or RNASEL deficiency caused by biallelic loss-of-function or hypomorphic variants were identified. Variants in these genes were absent from a control group of 1288 patients with mild or asymptomatic infection and a group of 159 children with COVID-19 pneumonia. Interestingly, these variants are estimated to be present in homozygous form in 1 in 10,000 individuals in the general population, which is consistent with the prevalence of MIS-C. As such, AR OAS1, OAS2, or RNASEL deficiency could explain about 1% of MIS-C cases globally. The authors of this same study36 show that patients’ fibroblasts and gene-edited epithelial cell lines lacking OAS1, OAS2, or RNASEL are not more susceptible to SARS-CoV-2 infection, nor are OAS1-, OAS2-, or RNASEL-deleted monocytes more permissive to viral replication. On the other hand, both gene-targeted monocytes and primary monocytes from patients display an exaggerated inflammatory response to dsRNA or SARS-CoV-2 infection, with hyperproduction of several proinflammatory cytokines, such as IL-6, CXCL9, CXCL10, and TNF (the levels of which are also elevated in the serum of patients with MIS-C), and with an upregulated proinflammatory gene expression profile at the transcriptomic level. Thus, in the absence of functional OAS-RNase L signaling, hyperinflammation is driven by activation of the RIG-I and melanoma differentiation-associated protein 5 (MDA5)–mitochondrial antiviral signaling (MAVS) pathway in response to cytosolic dsRNA sensing in monocytes. The reason why there is typically a delay of several weeks between the original infection and the onset of hyperinflammation in children with MIS-C currently remains unexplained.36
Finally, several other groups have screened cohorts of children with MIS-C by means of exome sequencing or targeted panels and have found enrichment in rare variants in genes related to autoimmunity, autoinflammation, and immune dysregulation, including genes underlying HLH (LYST, STXBP2, UNC13D, PRF1, AP3B1, and DOCK8) and genes involved in the interferon responses (IFNB1, IFNA21, IFNA4, IFNA6, IFIH1, TLR3, TRAF3, IRF3, IFNAR1, and IFNAR2); however, most of these studies do not functionally validate the pathogenicity of the variants and the correlation therefore remains only hypothetical.18 , 91, 92, 93
Discussion
The emergence of SARS-CoV-2 in December 2019 and into early 2020 has had a devastating impact worldwide. However, it has also provided the rare opportunity of studying a novel pathogen in a completely naive population. As expected from our knowledge of the human genetics of critical influenza, type I interferons are central to the human immunologic defense in COVID-19.
A defective type I interferon response in the first phases of the viral infection correlates with more severe disease and sustained viremia driving hyperinflammation and multiorgan involvement at a later stage.94 A 2-phase pathophysiologic model has been proposed; according to this model, uncontrolled viral infection due to a defective type I interferon response in several inborn errors of interferon immunity is followed by hyperactivation and recruitment of leukocytes, which ultimately lead to excessive inflammation.36 , 38 , 86, 95, 96 Studies investigating the genetic and immunologic determinants of critical COVID-19 have confirmed the crucial role of type I interferon immunity by revealing inborn errors of type I interferon immunity and their autoimmune phenocopies underlying critical or fatal COVID-19 (Tables I and II), as well as common polymorphisms conferring a higher risk of severe infection. Some of these defects had previously been shown to underlie critical influenza pneumonia (TLR3 and IRF7) or other severe viral infections (IFNAR1, IFNAR2, STAT2, IRF3, TBK1, UNC93B1, and TRIF), whereas defects of TLR7 were discovered as a novel cause of x-linked recessive severe COVID-19 pneumonia.15 , 16 , 18, 19, 20, 21 , 25 These examples highlight the redundancy of most of these mediators of type I interferon immunity in as much as pathogenic loss-of-function variants confer susceptibility to a very narrow spectrum of viruses. Thus, TLR7 seems redundant in human defense against influenza but necessary for defense against COVID-19. Similarly, neutralizing autoantibodies against type I interferon are responsible for up to 20% of critical and fatal COVID-19 cases and were found in only 5% of patients with critical influenza pneumonia.28 , 29 , 79 T-cell defects typically predispose to severe infection with a broad range of viruses and opportunistic pathogens, so an increased morbidity and mortality would be expected in patients with combined immune defects. This is confirmed by the finding of an extremely elevated fatality rate in children with severe combined immunodeficiency before transplantation, whereas survival was 100% in children who were infected with SARS-CoV-2 after curative procedures.14 , 98, 99, 100, 101, 102 Very few severe or lethal cases have been reported in patients with combined immunodeficiencies other than severe combined immunodeficiency, possibly indicating a subtle role of T-cell immunity in clearing SARS-CoV-2 compared with the robust contribution of type I interferon responses.39 On the other hand, the pathophysiologic mechanism underlying MIS-C seems to be an exacerbated inflammatory process in which the type I and II interferons, IL-6, and RIG-I-MDA5-MAVS pathways play a central role and the monocytes are the key drivers of inflammation, as confirmed by reports of several inborn errors of immune dysregulation (defects of SOCS1, XIAP, and CYBB) and RNA degradation (defects of OAS1, OAS2, and RNASEL) in children with MIS-C.34, 35, 36
On the basis of these findings, attempts to devise a therapeutic strategy based on modulating the interferon response have been proposed.37 A very fine balance between initial activation of antiviral and inflammatory responses to control the viral spread and subsequent downtuning of the inflammation to avoid organ damage seems to be required. The in-depth study of patients with critical COVID-19 in terms of genetic and immunologic determinants has undoubtedly expanded the awareness of the genetic (and immunologic) theory of infectious diseases beyond the realm of clinical immunology, reaching intensive care physicians and beyond. Novel inborn errors have been and will continue to be described in the context of critical COVID-19, furthering our knowledge and understanding of human immunology. Moreover, ongoing research will aid in understanding other manifestations of COVID-19, including long-COVID and neuro-COVID, which will in turn shed light on the postinfectious manifestations identified in other infections and their systemic and neurologic sequelae.
Acknowledgments
We wish to acknowledge all the doctors, nurses, caregivers, parents, and patient advocacy groups who have ensured the safety and well-being of patients with IEI during the COVID-19 pandemic. We want to thank Jean-Laurent Casanova and Helen Su for their leadership of the COVID Human Genetic Effort Consortium (www.covidhge.com) and all of the members listed at the end of this Acknowledgment for their insightful discussions and inspiration since the onset of the pandemic. We also wish to thank Stuart Tangye, Jean-Laurent Casanova, and the many members of the COVID Human Genetic Effort for providing constructive feedback on this review.
The members of the COVID Human Genetic Effort are as follows: Laurent Abel (Laboratory of Human Genetics of Infectious Diseases, Necker Branch, INSERM U1163, Necker Hospital for Sick Children, Paris, France; Paris Cité University, Imagine Institute, Paris, France; and St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, Rockefeller University, New York, NY), Salah Al-Muhsen (Immunology Research Lab, Department of Pediatrics, College of Medicine, King Saud University, Riyadh, Saudi Arabia), Alessandro Aiuti (San Raffaele Telethon Institute for Gene Therapy, IRCCS Ospedale San Raffaele, and Vita Salute San Raffaele University, Milan, Italy), Fahd Al-Mulla (Dasman Diabetes Institute, Department of Genetics and Bioinformatics, Dasman, Kuwait), Evangelos Andreakos (Laboratory of Immunobiology, Center for Clinical, Experimental Surgery and Translational Research, Biomedical Research Foundation of the Academy of Athens, Athens, Greece), Novelli Antonio (Laboratory of Medical Genetics, IRCCS Bambino Gesù Children’s Hospital, Rome, Italy), Andrés A. Arias (Group of Primary Immunodeficiencies, University of Antioquia UdeA, Medellin, Colombia), Sophie Trouillet-Assant (Hospices Civils de Lyon, International Center of Research in Infectiology, Lyon University, INSERM U1111, CNRS UMR 5308, ENS, UCBL, Lyon, France), Alexandre Belot (Pediatric Nephrology, Rheumatology, Dermatology, HFME, Hospices Civils de Lyon, National Referee Centre RAISE, and INSERM U1111, Université de Lyon, Lyon, France), Catherine M. Biggs (Department of Pediatrics, British Columbia Children’s Hospital, The University of British Columbia, Vancouver, British Columbia, Canada), Ahmed A. Bousfiha (Clinical Immunology Unit, Department of Pediatric Infectious Disease, CHU Ibn Rushd and the Laboratoire d’Immunologie Clinique, Inflammation et Allergie, Faculty of Medicine and Pharmacy, Hassan II University, Casablanca, Morocco), Alexandre Bolze (Helix, San Mateo, Calif), Alessandro Borghesi (Fondazione IRCCS Policlinico San Matteo di Pavia, Fellay Laboratory, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland), Petter Brodin (SciLifeLab, Department of Women’s and Children’s Health, Karolinska Institutet, Stockholm, Sweden), John Christodoulou (Murdoch Children’s Research Institute and Department of Paediatrics, University of Melbourne, Melbourne, Australia), Aurélie Cobat (Laboratory of Human Genetics of Infectious Diseases, Necker Branch, INSERM U1163, Necker Hospital for Sick Children, Paris, France; Paris Cité University, Imagine Institute, Paris, France; and St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, Rockefeller University, New York, NY), Antonio Condino-Neto (Department of Immunology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil), Stefan Constantinescu (de Duve Institute and Ludwig Cancer Research, Brussels, Belgium), Clifton L. Dalgard (Department of Anatomy, Physiology and Genetics, Uniformed Services University of the Health Sciences, Bethesda, Md), Xavier Marie Duval (INSERM CIC 1425 Paris, France; AP-HP, University Hospital of Bichat Paris, France, University Paris Diderot, Paris 7, UFR de Médecine-Bichat, Paris, France; IAME, INSERM, UMRS1137, University of Paris, Paris, France; and AP-HP, Bichat Claude Bernard Hospital, Infectious and Tropical Diseases Department, Paris, France), Sara Espinosa-Padilla (National Institute of Pediatrics, Mexico City, Mexico), Jacques Fellay (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, and Precision Medicine Unit, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland), Carlos Flores (Genomics Division, Instituto Tecnológico y de Energías Renovables, Santa Cruz de Tenerife, Spain; Research Unit, Hospital Universitario N.S. de Candelaria, Santa Cruz de Tenerife, Spain; CIBER de Enfermedades Respiratorias, Instituto de Salud Carlos III, Madrid, Spain; Department of Clinical Sciences, University Fernando Pessoa Canarias, Las Palmas de Gran Canaria, Spain), José Luis Franco (Group of Primary Immunodeficiencies, University of Antioquia UdeA, Medellin, Colombia), Antoine Froidure (Pulmonology Department, Cliniques Universitaires Saint-Luc, and Institut de Recherche Expérimentale et Clinique), Université Catholique de Louvain, Brussels, Belgium), Guy Gorochov (Sorbonne Université, Inserm, Centre d'Immunologie et des Maladies Infectieuses-Paris, Assistance Publique-Hôpitaux de Paris (AP-HP) Hôpital Pitié-Salpêtrière, Paris, France), Filomeen Haerynck (Department of Paediatric Immunology and Pulmonology, Centre for Primary Immunodeficiency Ghent, PID Research Laboratory, Jeffrey Modell Diagnosis and Research Centre, Ghent University Hospital, Ghent, Belgium), Rabih Halwani (Sharjah Institute of Medical Research, College of Medicine, University of Sharjah, Sharjah, United Arab Emirates), Elena W. Y. Hsieh (Departments of Pediatrics, Immunology and Microbiology, University of Colorado, School of Medicine, Aurora, Colo), Yuval Itan (Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, NY, and Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY), Erich D. Jarvis (Laboratory of Neurogenetics of Language, HHMI, The Rockefeller University, New York, NY), Kai Kisand (Molecular Pathology, Department of Biomedicine, Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu Estonia), Yu-Lung Lau (Department of Paediatrics and Adolescent Medicine, The University of Hong Kong, Hong Kong, China), Davood Mansouri (Department of Clinical Immunology and Infectious Diseases, National Research Institute of Tuberculosis and Lung Diseases, The Clinical Tuberculosis and Epidemiology Research Center, National Research Institute of Tuberculosis and Lung Diseases, Masih Daneshvari Hospital, Shahid Beheshti, University of Medical Sciences, Tehran, Iran) Isabelle Meyts (Department of Pediatrics, University Hospitals Leuven, and Department of Microbiology, Immunology and Transplantation, and Laboratory for Inborn Errors of Immunity, Katholieke Universiteit Leuven, Leuven, Belgium), Trine H. Mogensen (Department of Biomedicine, Aarhus University, Aarhus, Denmark), Lisa F. P. Ng (A∗STAR Infectious Disease Labs, Agency for Science, Technology and Research, Singapore, and Lee Kong Chian School of Medicine, Nanyang Technology University, Singapore), Luigi D. Notarangelo (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Md), Giuseppe Novelli (Department of Biomedicine and Prevention, Tor Vergata University of Rome, Rome, Italy), Satoshi Okada (Department of Pediatrics, Graduate School of Biomedical and Health Sciences, Hiroshima University, Hiroshima, Japan), Tayfun Ozcelik (Department of Molecular Biology and Genetics, Bilkent University, Bilkent, Ankara, Turkey), Rebeca Perez de Diego (Laboratory of Immunogenetics of Human Diseases, Innate Immunity Group, IdiPAZ Institute for Health Research, La Paz Hospital, Madrid, Spain), Carolina Prando (Faculdades Pequeno Príncipe, Instituto de Pesquisa Pelé Pequeno Príncipe, Curitiba, Brazil), Aurora Pujol (Neurometabolic Diseases Laboratory, Bellvitge Biomedical Research Institute, L’Hospitalet de Llobregat, Barcelona, Spain; Catalan Institution of Research and Advanced Studies, Barcelona, Spain, and Center for Biomedical Research on Rare Diseases, ISCIII, Barcelona, Spain), Lluis Quintana-Murci (Human Evolutionary Genetics Unit, CNRS U2000, Institut Pasteur, Paris, France, and Human Genomics and Evolution, Collège de France, Paris, France), Laurent Renia (A∗STAR Infectious Disease Labs, Agency for Science, Technology and Research, Singapore; Lee Kong Chian School of Medicine, Nanyang Technology University, Singapore), Igor Resnick (Department of Medical Genetics, Medical University, Varna and Department Hematology and BMT, University Hospital St. Marina, Bulgaria), Lucie Roussel (Department of Medicine, Division of Infectious Diseases, McGill University Health Centre, Montréal, Québec, Canada, and Infectious Disease Susceptibility Program, Research Institute, McGill University Health Centre, Montréal, Québec, Canada), Carlos Rodríguez-Gallego (Department of Immunology, University Hospital of Gran Canaria Dr. Negrín, Canarian Health System, Las Palmas de Gran Canaria, Spain; Department of Medical and Surgical Sciences, School of Medicine, University of Las Palmas de Gran Canaria, Las Palmas de Gran Canaria, Spain, and Department of Clinical Sciences, University Fernando Pessoa Canarias, Las Palmas de Gran Canaria, Spain), Vanessa Sancho-Shimizu (Department of Paediatric Infectious Diseases and Virology, Imperial College London, London, UK, and Centre for Paediatrics and Child Health, Faculty of Medicine, Imperial College London, London, UK), Mohammad Shahrooei (Specialized Immunology Laboratory, Ahvaz, Iran; Department of Microbiology and Immunology, Clinical and Diagnostic Immunology, Katholieke Universiteit Leuven, Leuven, Belgium), Pere Soler-Palacín (Pediatric Infectious Diseases and Immunodeficiencies Unit, Vall d’Hebron Barcelona Hospital Campus, Barcelona, Spain), András N. Spaan (St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY, and Department of Medical Microbiology, University Medical Center Utrecht, Utrecht, The Netherlands), Ivan Tancevski (Department of Internal Medicine II, Medical University of Innsbruck, Innsbruck, Austria), Stuart G. Tangye (Garvan Institute of Medical Research, Darlinghurst, New South Wales, Australia, and St. Vincent’s Clinical School, Faculty of Medicine, University of New South Wales, Sydney, New South Wales, Australia), Ahmad Abou Tayoun (Al Jalila Children’s Hospital, Dubai, United Arab Emerates), Şehime Gülsün Temel (Department of Medical Genetics, Department of Histology and Embryology, Faculty of Medicine, Bursa Uludağ University, Bursa, Turkey; and Department of Translational Medicine, Health Sciences Institute, Bursa Uludağ University, Bursa, Turkey), Pierre Tiberghien (Etablissement Francais Du Sang, La Plaine-Saint Denis, Saint-Denis, France) Jordi Perez Tur (Institut de Biomedicina de València-CSIC, CIBERNED, Unitat Mixta de Neurologia i Genètica, IIS La Fe, Vallencia, Spain), Stuart E. Turvey (BC Children’s Hospital, The University of British Columbia, Vancouver, Canada), Furkan Uddin (Centre for Precision Therapeutics, Genetics and Genomic Medicine Centre, Holy Family Red Crescent Medical College Dhaka, Bangladesh) Mohammed J. Uddin (College of Medicine, Mohammed Bin Rashid University of Medicine and Health Sciences, Dubai, United Arab Emirates; Cellular Intelligence Lab, GenomeArc, Inc, Toronto, Ontario, Canada), Mateus Vidigal (University of São Paulo, Brazil), Donald C. Vinh (Department of Medicine, Division of Infectious Diseases, McGill University Health Centre, Montréal, Québec, Canada, and Infectious Disease Susceptibility Program, Research Institute, McGill University Health Centre, Montréal, Québec, Canada), Mayana Zatz (Biosciences Institute, University of São Paulo, São Paulo, Brazil), Keisuke Okamoto (Tokyo Medical and Dental University, Tokyo, Japan), David S. Perlin (Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, NJ), Graziano Pesole (Department of Biosciences, Biotechnology and Biopharmaceutics, University of Bari A. Moro, Bari, Italy), Christian Thorball (Precision Medicine Unit, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland.) Diederik van de Beek (Department of Neurology, Amsterdam Neuroscience, Amsterdam University Medical Center, University of Amsterdam, Amsterdam, The Netherlands), Roger Colobran (Hospital Universitari Vall d’Hebron, Barcelona, Spain), Joost Wauters (Department of General Internal Medicine, Medical Intensive Care Unit, University Hospitals Leuven, Leuven, Belgium), Shen-Ying Zhang (Laboratory of Human Genetics of Infectious Diseases, Necker Branch, INSERM U1163, Necker Hospital for Sick Children, Paris, France; Paris Cité University, Imagine Institute, Paris, France; and St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, Rockefeller University, New York, NY), Qian Zhang(Laboratory of Human Genetics of Infectious Diseases, Necker Branch, INSERM U1163, Necker Hospital for Sick Children, Paris, France; Paris Cité University, Imagine Institute, Paris, France; and St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, Rockefeller University, New York, NY), Helen C. Su (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Md), and Jean-Laurent Casanova (The Rockefeller University and Howard Hughes Medical Institute, New York, NY, and Necker Hospital for Sick Children and INSERM, Paris, France).
Footnotes
Supported by the European Reference Network for Rare Immunodeficiency, Autoinflammatory and Autoimmune Diseases (ERN-RITA). I.M. is a senior clinical investigator at the Research Foundation–Flanders (FWO Vlaanderen) and is supported by a Katholieke Universiteit Leuven C1 (grant C16/18/007), the European Commission under Horizon Europe project UNDINE GA 10127100, the Research Foundation–Flanders (grants G0C8517N, G0B5120N, and G0E8420N), a European Research Council Starting Grant, and the Jeffrey Modell Foundation. H.C.S. and L.D.N. (listed under the COVID-Human Genetic Effort Consortium Consortium) are supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, NIH. J.L.C. is supported by the National Institutes of Health (grants R01AI088364, R01AI163029, and UL1TR001866), Fisher Center for Alzheimer’s Research Foundation, Meyer Foundation, JPB Foundation, French National Research Agency (grants ANR-10-IAHU-01, ANR-10-LABX-62-IBEID, ANR-20-CE93-003, ANR-20-CO11-0001), French Foundation for Medical Research (grant EQU201903007798), ANRS-COV05, the European Union’s Horizon 2020 Research and Innovation Program (grant 824110; EASI-Genomics), HORIZON-HLTH-2021-DISEASE-04 Program (grant 01057100; UNDINE), the ANR-RHU COVIFERON Program (grant ANR-21-RHUS-08), the Square Foundation, Grandir-Fonds de solidarité pour l’enfance, the Fondation du Souffle, the SCOR Corporate Foundation for Science, and the French Ministry of Higher Education, Research, and Innovation (grant MESRI-COVID-19).
Disclosure of potential conflict of interest: The authors declare that they have no conflicts of interest relevant to this manuscript.
Contributor Information
COVID Human Genetic Effort:
Laurent Abel, Salah Al-Muhsen, Alessandro Aiuti, Fahd Al-Mulla, Evangelos Andreakos, Novelli Antonio, Andrés A. Arias, Sophie Trouillet-Assant, Alexandre Belot, Catherine M. Biggs, Ahmed A. Bousfiha, Alex Bolze, Alessandro Borghesi, Petter Brodin, John Christodoulou, Aurélie Cobat, Antonio Condino-Neto, Stefan Constantinescu, Clifton L. Dalgard, Sara Espinosa-Padilla, Jacques Fellay, Carlos Flores, José Luis Franco, Antoine Froidure, Guy Gorochov, Filomeen Haerynck, Rabih Halwani, Elena W.Y. Hsieh, Yuval Itan, Kai Kisand, Yu-Lung Lau, Davood Mansouri, Isabelle Meyts, Trine H. Mogensen, Lisa F.P. Ng, Luigi D. Notarangelo, Giuseppe Novelli, Satoshi Okada, Tayfun Ozcelik, Rebeca Perez de Diego, Carolina Prando, Aurora Pujol, Lluis Quintana-Murci, Laurent Renia, Igor Resnick, Lucie Roussel, Carlos Rodríguez-Gallego, Vanessa Sancho-Shimizu, Mohammed Shahrooei, Pere Soler-Palacín, András N. Spaan, Ivan Tancevski, Stuart G. Tangye, Ahmad Abou Tayoun, Şehime Gülsün Temel, Pierre Tiberghien, Jordi Perez Tur, Stuart E. Turvey, Furkan Uddin, Mohammed J. Uddin, Mateus Vidigal, Donald C. Vinh, Mayana Zatz, Keisuke Okamoto, David S. Perlin, Graziano Pesole, Christian Thorball, Diederik van de Beek, Roger Colobran, Joost Wauters, Shen-Ying Zhang, Qian Zhang, Helen C. Su, and Jean-Laurent Casanova
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early Transmission dynamics in wuhan, china, of novel coronavirus–infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasselli G., Zangrillo A., Zanella A., Antonelli M., Cabrini L., Castelli A., et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region. Italy. JAMA. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucciol G., Moens L., Bosch B., Bossuyt X., Casanova J.L., Puel A., et al. Lessons learned from the study of human inborn errors of innate immunity. J Allergy Clin Immunol. 2019;143:507–527. doi: 10.1016/j.jaci.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond M.S., Kanneganti T.D. Innate immunity: the first line of defense against SARS-CoV-2. Nat Immunol. 2022;23:165–176. doi: 10.1038/s41590-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long B., Carius B.M., Chavez S., Liang S.Y., Brady W.J., Koyfman A., et al. Clinical update on COVID-19 for the emergency clinician: presentation and evaluation. Am J Emerg Med. 2022;54:46–57. doi: 10.1016/j.ajem.2022.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Driscoll M., Ribeiro Dos Santos G., Wang L., Cummings D.A.T., Azman A.S., Paireau J., et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590:140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 9.Casanova J.L., Su H.C., Abel L., Aiuti A., Almuhsen S., Arias A.A., et al. A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell. 2020;181:1194–1199. doi: 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niemi M.E.K., Karjalainen J., Liao R.G., Neale B.M., Daly M., Ganna A., et al. Mapping the human genetic architecture of COVID-19. Nature. 2021;600:472–477. doi: 10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 12.Severe Covid-19 GWAS Group. Aghemo A., Angelini C., Badalamenti S., Balzarini L., Bocciolone M., et al. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shelton J.F., Shastri A.J., Ye C., Weldon C.H., Filshtein-Sonmez T., Coker D., et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat Genet. 2021;53:801–808. doi: 10.1038/s41588-021-00854-7. [DOI] [PubMed] [Google Scholar]
- 14.Meyts I., Bucciol G., Quinti I., Neven B., Fischer A., Seoane E., et al. Coronavirus disease 2019 in patients with inborn errors of immunity: an international study. J Allergy Clin Immunol. 2021;147:520–531. doi: 10.1016/j.jaci.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q., Bastard P., Liu Z., Pen J.L., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell T.M., Liu Z., Zhang Q., Moncada-Velez M., Covill L.E., Zhang P., et al. Respiratory viral infections in otherwise healthy humans with inherited IRF7 deficiency. J Exp Med. 2022;219 doi: 10.1084/jem.20220202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciancanelli M.J., Huang S.X.L., Luthra P., Garner H., Itan Y., Volpi S., et al. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348:448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abolhassani H., Landegren N., Bastard P., Materna M., Modaresi M., Du L., et al. Inherited IFNAR1 deficiency in a child with both critical COVID-19 pneumonia and multisystem inflammatory syndrome. J Clin Immunol. 2022;42:471–483. doi: 10.1007/s10875-022-01215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khanmohammadi S., Rezaei N., Khazaei M., Shirkani A. A case of autosomal recessive interferon alpha/beta receptor alpha chain (IFNAR1) deficiency with severe COVID-19. J Clin Immunol. 2022;42:19–24. doi: 10.1007/s10875-021-01166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q., Matuozzo D., Le Pen J., Lee D., Moens L., Asano T., et al. Recessive inborn errors of type I IFN immunity in children with COVID-19 pneumonia. J Exp Med. 2022;219 doi: 10.1084/jem.20220131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asano T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Renkilaraj M., et al. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solanich X., Vargas-Parra G., van der Made C.I., Simons A., Schuurs-Hoeijmakers J., Antolí A., et al. Genetic screening for TLR7 variants in young and previously healthy men with severe COVID-19. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.719115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324:663. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim H.K., Huang S.X.L., Chen J., Kerner G., Gilliaux O., Bastard P., et al. Severe influenza pneumonitis in children with inherited TLR3 deficiency. J Exp Med. 2019;216:2038–2056. doi: 10.1084/jem.20181621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt A., Peters S., Knaus A., Sabir H., Hamsen F., Maj C., et al. TBK1 and TNFRSF13B mutations and an autoinflammatory disease in a child with lethal COVID-19. NPJ Genom Med. 2021;6:55. doi: 10.1038/s41525-021-00220-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahmood H.Z., Madhavarapu S., Almuqamam M. Varying illness severity in patients with MyD88 deficiency infected with coronavirus SARS-CoV-2 [meeting abstract] Pediatrics. 2021;147:453–454. [Google Scholar]
- 27.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bastard P., Orlova E., Sozaeva L., Lévy R., James A., Schmitt M.M., et al. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J Exp Med. 2021;218 doi: 10.1084/jem.20210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., et al. Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years old and account for ∼20% of COVID-19 deaths. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manry J., Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., et al. The risk of COVID-19 death is much greater and age dependent with type I IFN autoantibodies. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2200413119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bastard P., Vazquez S., Liu J., Laurie M.T., Wang C.Y., Gervais A., et al. Vaccine breakthrough hypoxemic COVID-19 pneumonia in patients with auto-Abs neutralizing type I IFNs. Sci Immunol. 2022;0 doi: 10.1126/sciimmunol.abp8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q., Bastard P., COVID Human Genetic Effort. Karbuz A., Gervais A., Tayoun A.A., et al. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature. 2022;603:587–598. doi: 10.1038/s41586-022-04447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., et al. Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years old and account for ∼20% of COVID-19 deaths. Sci Immunol. 2021;6(62) doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou J., Platt C.D., Habiballah S., Nguyen A.A., Elkins M., Weeks S., et al. Mechanisms underlying genetic susceptibility to multisystem inflammatory syndrome in children (MIS-C) J Allergy Clin Immunol. 2021;142:732–738. doi: 10.1016/j.jaci.2021.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee P.Y., Platt C.D., Weeks S., Grace R.F., Maher G., Gauthier K., et al. Immune dysregulation and multisystem inflammatory syndrome in children (MIS-C) in individuals with haploinsufficiency of SOCS1. J Allergy Clin Immunol. 2020;146:1194–1200.e1. doi: 10.1016/j.jaci.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee D., Le Pen J., Yatim A., Dong B., Aquino Y., Ogishi M., et al. Inborn errors of OAS-RNase L in SARS-CoV-2-related multisystem inflammatory syndrome in children. Science. 2022;0 doi: 10.1126/science.abo3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vinh D.C., Abel L., Bastard P., Cheng M.P., Condino-Neto A., Gregersen P.K., et al. Harnessing type I interferon immunity against SARS-CoV-2 with early administration of IFN-β. J Clin Immunol. 2021;41:1425–1442. doi: 10.1007/s10875-021-01068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q., Bastard P., Bolze A., Jouanguy E., Zhang S.Y., Cobat A., et al. Life-threatening COVID-19: defective interferons unleash excessive inflammation. Med (N Y) 2020;1:14–20. doi: 10.1016/j.medj.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bucciol G., Tangye S.G., Meyts I. Coronavirus disease 2019 in patients with inborn errors of immunity: lessons learned. Curr Opin Pediatr. 2021;33:648–656. doi: 10.1097/MOP.0000000000001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernandez N., Bucciol G., Moens L., Le Pen J., Shahrooei M., Goudouris E., et al. Inherited IFNAR1 deficiency in otherwise healthy patients with adverse reaction to measles and yellow fever live vaccines. J Exp Med. 2019;216:2057–2070. doi: 10.1084/jem.20182295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gothe F., Hatton C.F., Truong L., Klimova Z., Kanderova V., Fejtkova M., et al. A novel case of homozygous interferon alpha/beta receptor alpha chain (IFNAR1) deficiency with hemophagocytic lymphohistiocytosis. Clin Infect Dis. 2020;74:136–139. doi: 10.1093/cid/ciaa1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bastard P., Manry J., Chen J., Rosain J., Seeleuthner Y., AbuZaitun O., et al. Herpes simplex encephalitis in a patient with a distinctive form of inherited IFNAR1 deficiency. J Clin Invest. 2021;131 doi: 10.1172/JCI139980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bastard P., Hsiao K.C., Zhang Q., Choin J., Best E., Chen J., et al. A loss-of-function IFNAR1 allele in Polynesia underlies severe viral diseases in homozygotes. J Exp Med. 2022;219 doi: 10.1084/jem.20220028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duncan C.J.A., Mohamad S.M.B., Young D.F., Skelton A.J., Leahy T.R., Munday D.C., et al. Human IFNAR2 deficiency: lessons for antiviral immunity. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aac4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duncan C.J.A., Skouboe M.K., Howarth S., Hollensen A.K., Chen R., Børresen M.L., et al. Life-threatening viral disease in a novel form of autosomal recessive IFNAR2 deficiency in the Arctic. J Exp Med. 2022;219 doi: 10.1084/jem.20212427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bucciol G., Moens L., Corveleyn A., Dreesman A., Meyts I. A novel kindred with MyD88 deficiency. J Clin Immunol. 2022:1–4. doi: 10.1007/s10875-022-01240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.García-García A, Pérez de Diego R, Flores C, Rinchai D, Solé-Violán J, Àngela Deyà-Martínez, et al. Humans with inherited MyD88 and IRAK-4 deficiencies are predisposed to hypoxemic COVID-19 pneumonia. J Exp Med, 10.1084/jem.20220170. [DOI] [PMC free article] [PubMed]
- 48.Sheahan T., Morrison T.E., Funkhouser W., Uematsu S., Akira S., Baric R.S., et al. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLOS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Onodi F., Bonnet-Madin L., Meertens L., Karpf L., Poirot J., Zhang S.Y., et al. SARS-CoV-2 induces human plasmacytoid predendritic cell diversification via UNC93B and IRAK4 SARS-CoV-2 and plasmacytoid predendritic cells. J Exp Med. 2021;218 doi: 10.1084/jem.20201387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matuozzo D, Talouarn E, Marchal A, Manry J, Seeleuthner Y, Zhang Y, et al. Rare predicted loss-of-function variants of type I IFN immunity genes are associated with life-threatening COVID-19 2022;2022.10.22.22281221. [DOI] [PMC free article] [PubMed]
- 51.Kousathanas A., Pairo-Castineira E., Rawlik K., Stuckey A., Odhams C.A., Walker S., et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature. 2022;607:97–103. doi: 10.1038/s41586-022-04576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Casanova J.L., Abel L. From rare disorders of immunity to common determinants of infection: Following the mechanistic thread. Cell. 2022;185:3086–3103. doi: 10.1016/j.cell.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Döffinger R., Helbert M.R., Barcenas-Morales G., Yang K., Dupuis S., Ceron-Gutierrez L., et al. Autoantibodies to interferon-γ in a patient with selective susceptibility to mycobacterial infection and organ-specific autoimmunity. Clin Infect Dis. 2004;38:e10–e14. doi: 10.1086/380453. [DOI] [PubMed] [Google Scholar]
- 54.Puel A., Picard C., Lorrot M., Pons C., Chrabieh M., Lorenzo L., et al. Recurrent staphylococcal cellulitis and subcutaneous abscesses in a child with autoantibodies against IL-6. J Immunol. 2008;180:647–654. doi: 10.4049/jimmunol.180.1.647. [DOI] [PubMed] [Google Scholar]
- 55.Kisand K., Bøe Wolff A.S., Podkrajšek K.T., Tserel L., Link M., Kisand K.V., et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J Exp Med. 2010;207:299–308. doi: 10.1084/jem.20091669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ku C.L., Chi C.Y., von Bernuth H., Doffinger R. Autoantibodies against cytokines: phenocopies of primary immunodeficiencies? Hum Genet. 2020;139:783–794. doi: 10.1007/s00439-020-02180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puel A., Döffinger R., Natividad A., Chrabieh M., Barcenas-Morales G., Picard C., et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med. 2010;207:291–297. doi: 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vallbracht A., Treuner J., Flehmig B., Joester K.E., Niethammer D. Interferon-neutralizing antibodies in a patient treated with human fibroblast interferon. Nature. 1981;289:496–497. doi: 10.1038/289496a0. [DOI] [PubMed] [Google Scholar]
- 59.Panem S., Check I.J., Henriksen D., Vilcek J. Antibodies to alpha-interferon in a patient with systemic lupus erythematosus. J Immunol. 1982;129:1–3. [PubMed] [Google Scholar]
- 60.Walter J.E., Rosen L.B., Csomos K., Rosenberg J.M., Mathew D., Keszei M., et al. Broad-spectrum antibodies against self-antigens and cytokines in RAG deficiency. J Clin Invest. 2015;125:4135–4148. doi: 10.1172/JCI80477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levin M. Anti-interferon auto-antibodies in autoimmune polyendocrinopathy syndrome type 1. PLOS Med. 2006 giu;3:e292. doi: 10.1371/journal.pmed.0030292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eriksson D., Bacchetta R., Gunnarsson H.I., Chan A., Barzaghi F., Ehl S., et al. The autoimmune targets in IPEX are dominated by gut epithelial proteins. J Allergy Clin Immunol. 2019;144:327–330.e8. doi: 10.1016/j.jaci.2019.02.031. [DOI] [PubMed] [Google Scholar]
- 63.Shiono H., Wong Y.L., Matthews I., Liu J., Zhang W., Sims G., et al. Spontaneous production of anti-IFN-α and anti-IL-12 autoantibodies by thymoma cells from myasthenia gravis patients suggests autoimmunization in the tumor. Int Immunol. 2003;15:903–913. doi: 10.1093/intimm/dxg088. [DOI] [PubMed] [Google Scholar]
- 64.Rosenberg J.M., Maccari M.E., Barzaghi F., Allenspach E.J., Pignata C., Weber G., et al. Neutralizing anti-cytokine autoantibodies against interferon-α in immunodysregulation polyendocrinopathy enteropathy X-linked. Front Immunol. 2018;9:544. doi: 10.3389/fimmu.2018.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pozzetto B., Mogensen K.E., Tovey M.G., Gresser I. Characteristics of autoantibodies to human interferon in a patient with varicella-zoster disease. J Infect Dis. 1984;150:707–713. doi: 10.1093/infdis/150.5.707. [DOI] [PubMed] [Google Scholar]
- 66.Mogensen K.E., Daubas P., Gresser I., Sereni D., Varet B. patient with circulating antibodies to α-interferon. Lancet. 1981;318:1227–1228. doi: 10.1016/s0140-6736(81)91460-4. [DOI] [PubMed] [Google Scholar]
- 67.Meyer S., Woodward M., Hertel C., Vlaicu P., Haque Y., Kärner J., et al. AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell. 2016;166:582–595. doi: 10.1016/j.cell.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beccuti G., Ghizzoni L., Cambria V., Codullo V., Sacchi P., Lovati E., et al. A COVID-19 pneumonia case report of autoimmune polyendocrine syndrome type 1 in Lombardy, Italy: letter to the editor. J Endocrinol Invest. 2020;43:1175–1177. doi: 10.1007/s40618-020-01323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van der Wijst M.G.P., Vazquez S.E., Hartoularos G.C., Bastard P., Grant T., Bueno R., et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simula E.R., Manca M.A., Noli M., Jasemi S., Ruberto S., Uzzau S., et al. Increased presence of antibodies against type i interferons and human endogenous retrovirus w in intensive care unit COVID-19 patients. Microbiol Spectr. 2022;10 doi: 10.1128/spectrum.01280-22. e01280-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathian A., Breillat P., Dorgham K., Bastard P., Charre C., Lhote R., et al. Lower disease activity but higher risk of severe COVID-19 and herpes zoster in patients with systemic lupus erythematosus with pre-existing autoantibodies neutralising IFN-α. Ann Rheum Dis. 2022;81:1695–1703. doi: 10.1136/ard-2022-222549. [DOI] [PubMed] [Google Scholar]
- 72.Frasca F., Scordio M., Santinelli L., Gabriele L., Gandini O., Criniti A., et al. Anti-IFN-α/-ω neutralizing antibodies from COVID-19 patients correlate with downregulation of IFN response and laboratory biomarkers of disease severity. Eur J Immunol. 2022;52:1120–1128. doi: 10.1002/eji.202249824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Akbil B., Meyer T., Stubbemann P., Thibeault C., Staudacher O., Niemeyer D., et al. Early and rapid identification of COVID-19 patients with neutralizing type I interferon auto-antibodies. J Clin Immunol. 2022;42:1111–1129. doi: 10.1007/s10875-022-01252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eto S., Nukui Y., Tsumura M., Nakagama Y., Kashimada K., Mizoguchi Y., et al. Neutralizing type I interferon autoantibodies in Japanese patients with severe COVID-19. J Clin Immunol. 2022:1–11. doi: 10.1007/s10875-022-01308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chauvineau-Grenier A., Bastard P., Servajean A., Gervais A., Rosain J., Jouanguy E., et al. Autoantibodies neutralizing type I interferons in 20% of COVID-19 deaths in a French hospital. J Clin Immunol. 2022;42:459–470. doi: 10.1007/s10875-021-01203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Busnadiego I., Abela I.A., Frey P.M., Hofmaenner D.A., Scheier T.C., Schuepbach R.A., et al. Critically ill COVID-19 patients with neutralizing autoantibodies against type I interferons have increased risk of herpesvirus disease. PLOS Biol. 2022;20 doi: 10.1371/journal.pbio.3001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ansari R., Rosen L.B., Lisco A., Gilden D., Holland S.M., Zerbe C.S., et al. Primary and acquired immunodeficiencies associated with severe varicella-zoster virus infections. Clin Infect Dis. 2021;73:e2705–e2712. doi: 10.1093/cid/ciaa1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hetemäki I., Laakso S., Välimaa H., Kleino I., Kekäläinen E., Mäkitie O., et al. Patients with autoimmune polyendocrine syndrome type 1 have an increased susceptibility to severe herpesvirus infections. Clin Immunol. 2021;231 doi: 10.1016/j.clim.2021.108851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Q., Pizzorno A., Miorin L., Bastard P., Gervais A., Le Voyer T., et al. Autoantibodies against type I IFNs in patients with critical influenza pneumonia. J Exp Med. 2022;219 doi: 10.1084/jem.20220514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bastard P., Michailidis E., Hoffmann H.H., Chbihi M., Le Voyer T., Rosain J., et al. Auto-antibodies to type I IFNs can underlie adverse reactions to yellow fever live attenuated vaccine. J Exp Med. 2021;218 doi: 10.1084/jem.20202486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verdoni L., Mazza A., Gervasoni A., Martelli L., Ruggeri M., Ciuffreda M., et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Toubiana J., Cohen J.F., Brice J., Poirault C., Bajolle F., Curtis W., et al. Distinctive features of Kawasaki disease following SARS-CoV-2 infection: a controlled study in Paris, France. J Clin Immunol. 2021;41:526–535. doi: 10.1007/s10875-020-00941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sancho-Shimizu V., Brodin P., Cobat A., Biggs C.M., Toubiana J., Lucas C.L., et al. SARS-CoV-2-related MIS-C: a key to the viral and genetic causes of Kawasaki disease? J Exp Med. 2021;218 doi: 10.1084/jem.20210446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gruber C.N., Patel R.S., Trachtman R., Lepow L., Amanat F., Krammer F., et al. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS-C) Cell. 2020;183:982–995.e14. doi: 10.1016/j.cell.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Feldstein L.R., Rose E.B., Horwitz S.M., Collins J.P., Newhams M.M., Son M.B.F., et al. Multisystem inflammatory syndrome in U.S. children and adolescents. N Engl J Med. 2020;383:334–346. doi: 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hadjadj J., Castro C.N., Tusseau M., Stolzenberg M.C., Mazerolles F., Aladjidi N., et al. Early-onset autoimmunity associated with SOCS1 haploinsufficiency. Nat Commun. 2020;11:5341. doi: 10.1038/s41467-020-18925-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mudde A.C.A., Booth C., Marsh R.A. Evolution of our understanding of XIAP Deficiency. Front Pediatr. 2021;9 doi: 10.3389/fped.2021.660520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Squire J.D., Vazquez S.N., Chan A., Smith M.E., Chellapandian D., Vose L., et al. Case report: secondary hemophagocytic lymphohistiocytosis with disseminated infection in chronic granulomatous disease—a serious cause of mortality. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.581475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schäppi M.G., Jaquet V., Belli D.C., Krause K.H. Hyperinflammation in chronic granulomatous disease and anti-inflammatory role of the phagocyte NADPH oxidase. Semin Immunopathol. 2008;30:255–271. doi: 10.1007/s00281-008-0119-2. [DOI] [PubMed] [Google Scholar]
- 91.Vagrecha A., Zhang M., Acharya S., Lozinsky S., Singer A., Levine C., et al. Hemophagocytic lymphohistiocytosis gene variants in multisystem inflammatory syndrome in children. Biology. 2022;11:417. doi: 10.3390/biology11030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Abuhammour W., Yavuz L., Jain R., Abu Hammour K., Al-Hammouri G.F., El Naofal M., et al. Genetic and clinical characteristics of patients in the middle east with multisystem inflammatory syndrome in children. JAMA Netw Open. 2022;5 doi: 10.1001/jamanetworkopen.2022.14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Santos-Rebouças C.B., Piergiorge R.M., dos Santos Ferreira C., Seixas Zeitel R. d.e., Gerber A.L., Rodrigues M.C.F., et al. Host genetic susceptibility underlying SARS-CoV-2-associated multisystem inflammatory syndrome in Brazilian children. Mol Med. 2022;28:153. doi: 10.1186/s10020-022-00583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Casanova J.L., Abel L. Mechanisms of viral inflammation and disease in humans. Science. 2021;374:1080–1086. doi: 10.1126/science.abj7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bordallo B., Bellas M., Cortez A.F., Vieira M., Pinheiro M. Severe COVID-19: what have we learned with the immunopathogenesis? Adv Rheumatol. 2020;60:50. doi: 10.1186/s42358-020-00151-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Castano-Jaramillo L.M., Yamazaki-Nakashimada M.A., O’Farrill-Romanillos P.M., Muzquiz Zermeño D., Scheffler Mendoza S.C., Venegas Montoya E., et al. COVID-19 in the context of inborn errors of immunity: a case series of 31 patients from Mexico. J Clin Immunol. 2021;41:1463–1478. doi: 10.1007/s10875-021-01077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goudouris E.S., Pinto-Mariz F., Mendonça L.O., Aranda C.S., Guimarães R.R., Kokron C., et al. Outcome of SARS-CoV-2 infection in 121 patients with inborn errors of immunity: a cross-sectional study. J Clin Immunol. 2021;81:1479–1489. doi: 10.1007/s10875-021-01066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marcus N., Frizinsky S., Hagin D., Ovadia A., Hanna S., Farkash M., et al. Minor clinical impact of COVID-19 pandemic on patients with primary immunodeficiency in Israel. Front Immunol. 2021;11 doi: 10.3389/fimmu.2020.614086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Keitel V., Bode J.G., Feldt T., Walker A., Müller L., Kunstein A., et al. Case report: convalescent plasma achieves SARS-CoV-2 viral clearance in a patient with persistently high viral replication over 8 weeks due to severe combined immunodeficiency (SCID) and graft failure. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.645989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Oers N.S.C., Hanners N.W., Sue P.K., Aquino V., Li Q.Z., Schoggins J.W., et al. SARS-CoV-2 infection associated with hepatitis in an infant with X-linked severe combined immunodeficiency. Clin Immunol. 2021;224 doi: 10.1016/j.clim.2020.108662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zamperlini-Netto G., Fernandes J.F., Garcia J.L., Ribeiro A.A.F., Camargo L.F.A., de Moraes Terra C., et al. COVID-19 after hematopoietic stem cell transplantation: report of two children. Bone Marrow Transplant. 2021;56:713–715. doi: 10.1038/s41409-020-01041-8. [DOI] [PubMed] [Google Scholar]