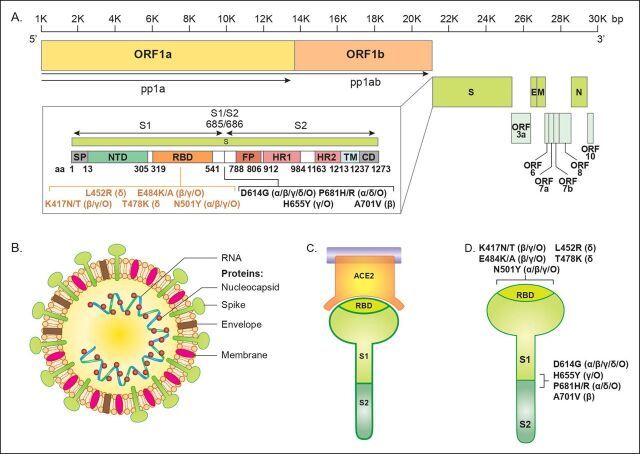
Figure 1.
Genome and structure of SARS-CoV-2. (A) SARS-CoV-2 genome and spike (S) protein amino acid composition. The SARS-CoV-2 genome is about 30 000 base pairs (bp) long and consists of open reading frames (ORF) and elements that are essential for the virus’ structure. The S protein is responsible for binding and entry into host cells. SARS-CoV-2 variants of concern contain various S protein non-synonymous mutations that result in amino acid changes in the receptor binding domain (orange bracketed text) and the S1/S2 subunit interface (black bracketed text), which have been shown to enhance transmissibility of the virus. Variants of concern include alpha (α), beta (β), gamma (γ), delta (δ), and omicron (O). (B) SARS-CoV-2 structure. SARS-CoV-2 is an RNA virus that has a crown-like appearance and contains four major structural proteins: nucleocapsid (N), spike (S), envelope (E), and membrane (M). (C) Viral S protein and human angiotensin converting enzyme 2 (ACE2) interaction. The SARS-CoV-2 S protein directly interacts with human ACE2 receptors in order to gain entry into host cells. The receptor binding domain (RBD) of the S protein tightly binds to ACE2. (D) S protein structure. The S protein protrudes out from the main SARS-CoV-2 bulk and is comprised of two subunits: S1 and S2. S1 contains the RBD, which directly interacts with the human ACE2 receptor, while the S1/S2 interface contains a furin cleavage site that is cleaved to allow S2 to fuse with the host cell membrane. Both the RBD and the S1/S2 interface contain transmissibility increasing mutations that are harboured in variants of concern