Figure 1.
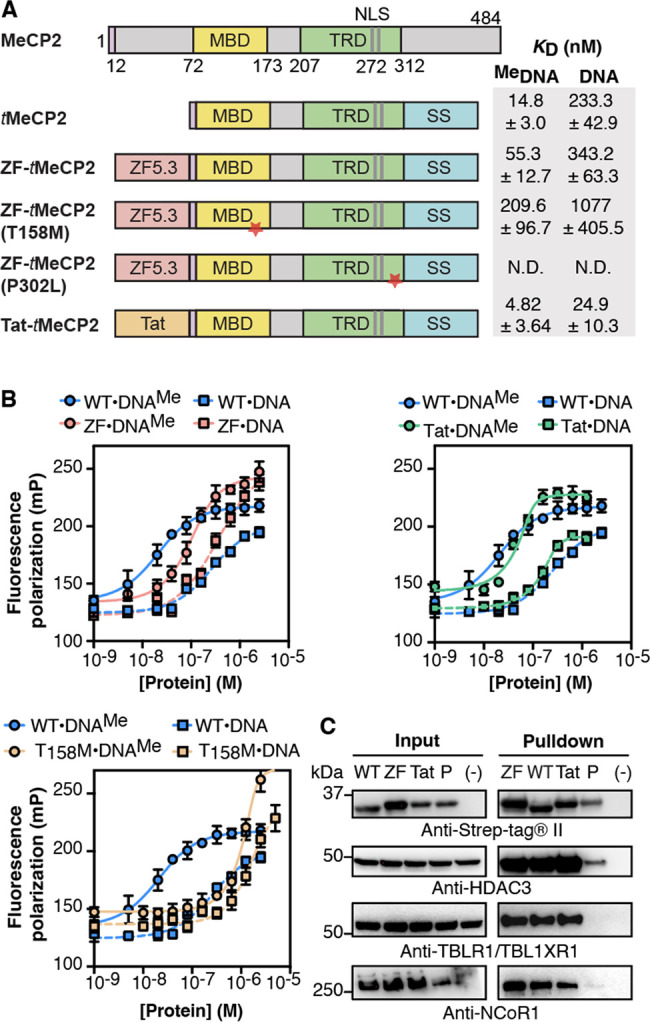
ZF-tMeCP2 is functional in vitro. (A) tMeCP2 proteins used in this work lack N-terminal residues 13–71 and C-terminal residues 313–484. All proteins were expressed in E. coli and purified as described in Supplementary Methods 1. We note that as the tMeCP2 proteins used in this work were expressed in bacteria, they lack post-translational arginine modifications that may affect heterochromatin accumulation, clustering and binding kinetics.50 MBD: methyl-CpG-binding domain; TRD: transcriptional-repressor domain; NLS: nuclear localization sequence; SS: sortase motif + Strep-tag. The red star indicates the location of the point mutation. (B) Plots showing changes in fluorescence polarization used to calculate the apparent equilibrium dissociation constant (KD) of the complex between each tMeCP2 variant and methylated (DNAMe) or nonmethylated (DNA) oligonucleotides (Table S5). The data were fitted to an equilibrium binding equation based on the Langmuir model51 to calculate the KD values in (A). Data are represented as mean ± SD. N.D.: not determined. (C) Western blots were used to analyze an in vitro anti-Strep-tag pull-down assay to probe the interaction of the indicated tMeCP2 variant (WT: tMeCP2, ZF: ZF-tMeCP2, Tat: Tat-tMeCP2, P: ZF-tMeCP2(P302L)) with subunits of the NCoR/SMRT repressor complex. HDAC3: Histone Deacetylase 3; TBLXR1: Transducin Beta-Like 1X-Related Protein 1; NCoR1: Nuclear Receptor Corepressor 1. The gel results shown are representative of three biological replicates.
