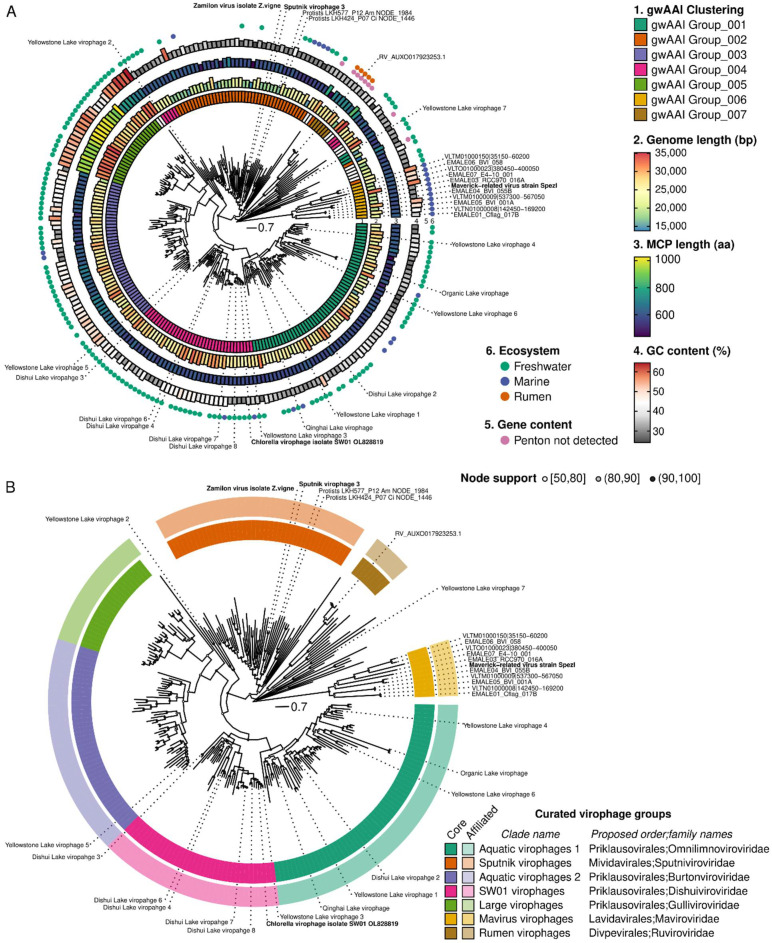
Figure 3.
Definition of new virophage clades based on MCP phylogeny and genome features. For both panels, an MCP phylogeny is presented based on complete and near-complete virophage genomes. For panel (A), genomes features are displayed including from the inner to the outer ring: (1) groups based on genome-wide amino-acid identity (gwAAI) considering the seven largest groups, (2) genome length, (3) Major Capsid Protein (MCP) length, (4) average GC content, (5) detection of the typical virophage penton protein based on HMM profiles, and (6) ecosystem from which the genome was obtained. In panel (B), the same tree is decorated with the classification of virophage genomes in curated groups. The “core” sequences (solid colors, inner ring) of each group represent the members of the monophyletic clade assigned to this group based on the gwAAI clustering (see panel (A)). The “affiliated” ring displays the results of a “best BLAST hit” affiliation of all sequences in the tree (ignoring self-hits) in light colors (outer ring).