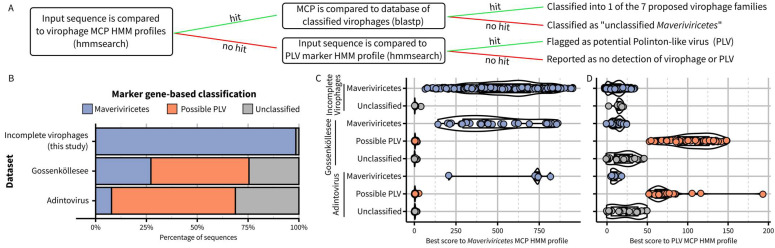
Figure 4.
Detection and classification of virophages from published datasets. (A) Schematic overview of the hmmsearch- and BLAST-based classification approach used to identify and assign virophages to the proposed families. For panels (B–D), three datasets were processed with the approach described in panel (A). The first includes non-redundant previously published virophage sequences gathered as part of this study and not included in the complete and near-complete genome set (n = 591; “Incomplete virophages”). The second includes contigs identified as virophages and Polinton-like viruses assembled from Gossenköllesee metagenomes and published in [19] (n = 114; “Gossenköllesee”). Finally, the third dataset included adintoviruses published in several previous studies [23,32,33] (n = 64; “Adintovirus”). In panel (B), the percentage of sequences assigned as Maveriviricetes (based on MCP HMM) or with a hit to the PLV HMM profile is indicated. The score distribution of score for the hits to the Maveriviricetes HMM or PLV HMM is displayed in panels (C,D), respectively, for each dataset and each group of sequence.