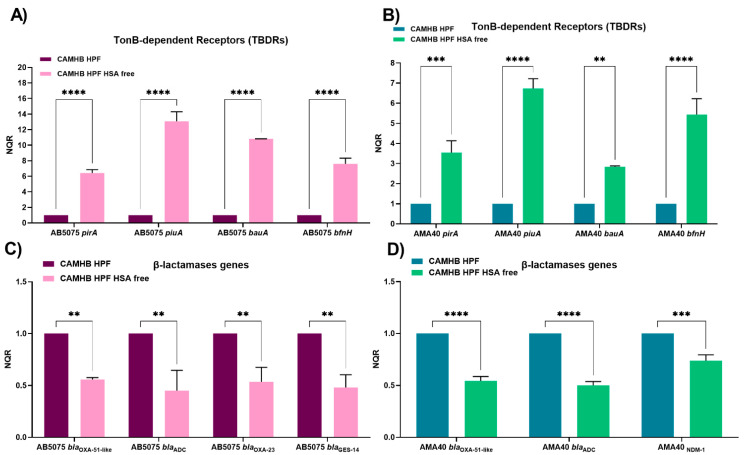
Figure 2.
Expression analysis of iron uptake genes and genes coding for β-lactamases in the AB5075 (A,C) and AMA40 (B,D) strains. The qRT-PCR of TonB-dependent receptors (pirA, piuA, bauA, and bfnH) and bla genes (blaOXA-51-like, blaADC, blaOXA-23, blaNDM-1, and blaGES-11) genes expressed in cation-adjusted Mueller Hinton (CAMHB) supplemented with either human pleural fluid (HPF) or HSA-free HPF. The data shown are the means ± SD of normalized relative quantities (NRQ) obtained from transcript levels calculated via the qBASE method. This method is a modification of the classic ΔΔCt method to take multiple reference genes (in this work, rpoB and recA) and gene-specific amplification efficiencies into account. At least three independent samples were used, and four technical replicates were performed from each sample. The CAMHB HPF was used as reference. Statistical significance (p < 0.05) was determined by two-way ANOVA followed by Tukey’s multiple comparison test, two asterisks: p < 0.01; three asterisks: p < 0.001; and four asterisks: p < 0.0001.