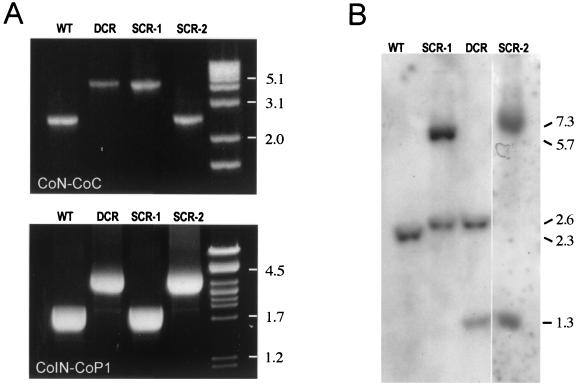
FIG. 4.
PCR mapping and Southern blot analysis of choE recombinants. (A) PCR mapping with CoN-CoC (upper panel) and CoIN-CoP1 (lower panel) primer pairs. See text and Fig. 3 legend for details. (B) Southern blot analysis. Genomic DNA from representative mutants of each recombinational type were cut with MluI and hybridized against the CoEN-CoEC probe covering the entire choE gene. Hybridization patterns of R. equi (WT), DCRs, and the two types of SCRs are shown. In R. equi, choE is flanked by two MluI sites separated by 2.3 kb; thus, the WT displays a single 2.3-kb hybridization band. pRHE3 has a single MluI site located at the middle of the aacC4 gene (Fig. 3); therefore, DCR shows two bands of 2.6 and 1.3 kb. In SCR-1, there were two bands of 2.6 and 5.7 kb. The 2.6-kb band is equivalent to the band of the same size observed in DCR and corresponds to the left side of the aacC4-disrupted choE allele; the 5.7-kb band includes the right side of the aacC4-disrupted choE allele, pUC19, and the intact copy of choE (Fig. 3). In SCR-2, two hybridization bands of 7.0 and 1.3 kb were observed (Fig. 3). When membranes were rehybridized with a pUC19 probe, hybridization signals were detected in SCR-1 and SCR-2, confirming the presence of the complete pRHE3 plasmid integrated at the R. equi choE locus as the result of an SCR event (data not shown).