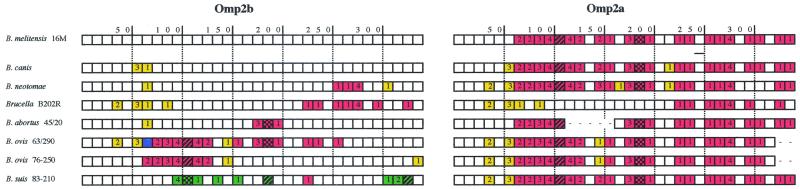
FIG. 4.
Schematic representation of multiple amino acid sequence alignment of the Omp2b size variants along with their corresponding Omp2a. This alignment includes sequences not obtained in this study but found in sequence databases. Sequences are subdivided in stretches of 10 residues, numbered following the B. melitensis 16M Omp2b sequence. A change in color indicates the presence of substitutions (the number inside the box indicates the number of substitutions compared to Omp2b of B. melitensis 16M among the 10 residues). The red color always indicates identity with motifs specific for Omp2a. The yellow color indicates sequence motifs not present in reference Omp2b nor in reference Omp2a but conserved between variants that share this color. The blue and green colors indicate motifs specific, respectively, of B. ovis 63/290 Omp2b and B. suis 83-210 Omp2b. Hatching in the box symbolizes a deletion; double hatching symbolizes an insertion, always compared to the Omp2b sequence of B. melitensis 16M. This figure clearly shows that motif exchanges between Omp2a and Omp2b are common among all Brucella strains, suggesting that gene conversion as a phenomenon occurred several times during Brucella evolution, thus explaining most of the Omp2b variation between strains, except the variation found in the B. suis 83-210 Omp2b porin. Interestingly, as shown in this figure, Omp2a of both B. ovis strains is truncated in its C-terminal end due to the presence of a stop codon, leading probably to a nonfunctional porin. B. abortus 45/20 Omp2a shows the same deletion as in B. abortus S19 Omp2a characterized by others (15, 16, 17, 25, 26).