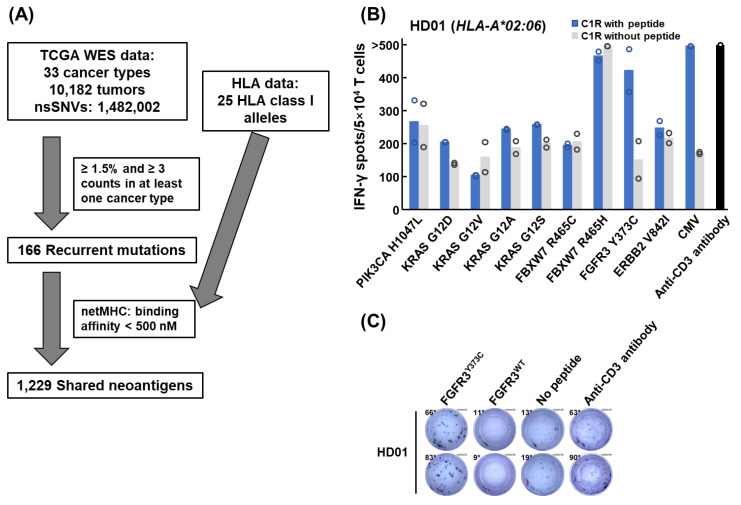
Figure 1.
Shared neoantigen screening from TCGA exome sequencing data. (A) Workflow for shared neoantigen screening. From 1,482,002 nonsynonymous single-nucleotide variants (nsSNVs) in 33 cancer types in TCGA exome data, 166 recurrent mutations were detected in more than 3 tumors, and 1.5% of tumors in at least one cancer type were extracted. Then, 1229 shared neoantigen peptides were predicted to be presented on at least one of 25 HLA class I molecules, which are frequently observed in Japanese populations. (B) In-vitro screening of shared neoantigen-reactive CD8+ T cells. IFN-γ ELISPOT screening of antigen-reactive CD8+ T cells for 9 shared neoantigen peptides using peripheral blood of healthy donor 1 (HD01) with HLA-A*02:06. Antigen-stimulated CD8+ T cells were co-cultured with C1R-A0206 cells pulsed with or without shared neoantigen peptides. Bars represented means in duplicate experiments (each circle indicates the number of spots). (C) Mutant FGFR3Y373C peptide-specific IFN-γ production of FGFR3Y373C-reactive CD8+ T cells. FGFR3Y373C-stimulated CD8+ T cells were co-cultured with C1R-A0206 cells pulsed with or without FGFR3Y373C or corresponding wild-type FGFR3WT peptide.