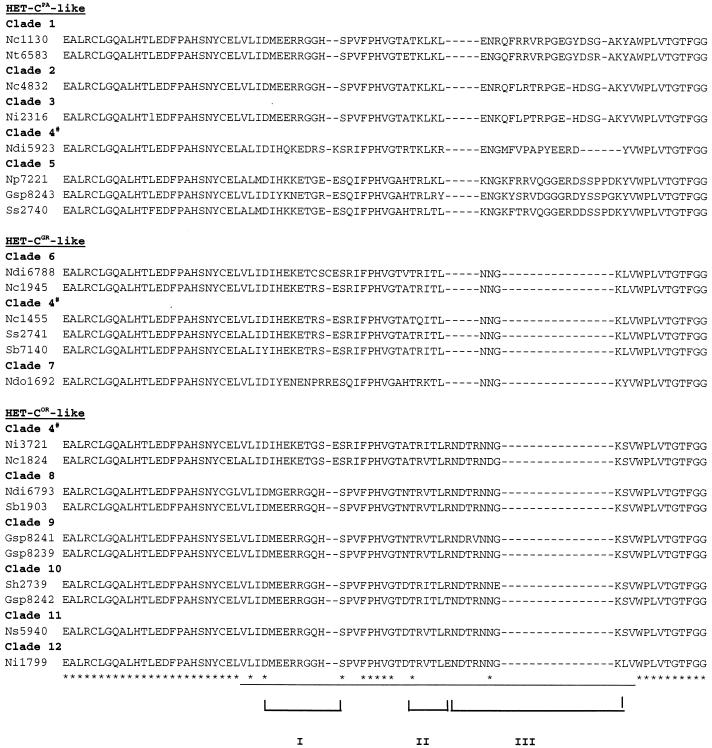
FIG. 2.
Predicted peptide sequences of the variable domain in naturally occurring het-c alleles. The underlined area is the het-c variable domain that is necessary and sufficient to confer het-c allelic specificity in the N. crassa het-cOR (FGSC 2489), het-cPA (FGSC 1130), and het-cGR (FGSC1945) alleles (39, 40). ∗, conserved sites. Dashes represent deletions. Regions I and II are polymorphic blocks; region III encompasses the indel motif. Each clade is supported by over 94% bootstrap values (48). HET-CPA-like, HET-COR-like, and HET-CGR-like refers to the indel pattern observed in the genetically characterized N. crassa het-cPA, het-cOR and het-cGR alleles, respectively (39). Clade 4 has representatives in all three HET-C-like classes based on the indel motif; separation of these three het-c types is supported by bootstrap values of less than 94%. GenBank accession numbers for nucleotide sequences of these alleles were previously cited (48); accession numbers are AF092695 to AF092732 and AF093679.