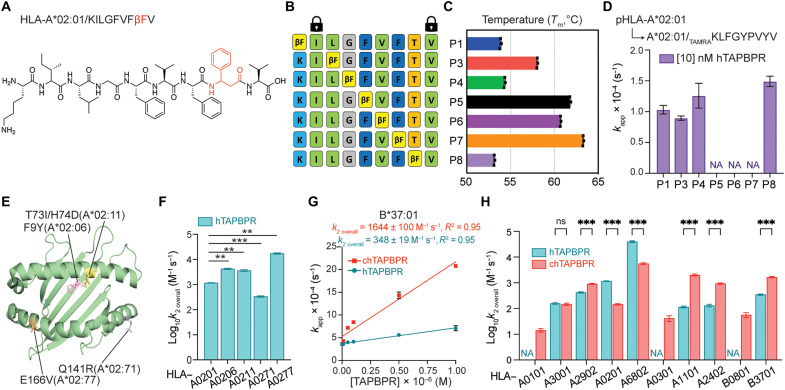
Fig. 4. Conditional peptide ligands reveal the effects of HLA micropolymorphisms and supertype dependence of peptide exchange by TAPBPR orthologs.
(A) Conditional peptide ligand KILGFVFβFV based on the HLA-A*02:01–restricted influenza matrix 1 (58–65, 78) epitope GILGFVFTL. T8 is replaced with βF. (B) HLA-A*02:01 βF scanning peptide panel at each position as indicated. (C) Tm (in degrees Celsius) values obtained from DSF of HLA-A*02:01 bound to KILGFVFTV with βF substitution at indicated positions. (D) Comparison of kapp for fluorescent peptide binding to HLA-A*02:01/KILGFVFTV with βF substitution at indicated positions in the presence of 10 nM hTAPBPR. (E) Crystal structure of HLA-A*02:01 (79) (PDB ID: 3MRE) indicates the polymorphic residues for HLA-A*02:06 (light pink), HLA-A*02:11 (light yellow), HLA-A*02:71 (light blue), and HLA-A*02:77 (light orange). (F) Log-scale comparison of k2overall for HLA-A*02:01, HLA-A*02:06, HLA-A*02:11, HLA-A*02:71, and HLA-A*02:77 in the presence of hTAPBPR. The two-sample unequal variance Student’s t test was performed, P > 0.12 (ns), *P < 0.033, **P < 0.002, and ***P < 0.001. (G) Linear correlations between the apparent rate constants kapp and the concentrations of TAPBPR orthologs for HLA-B*37:01. (H) Log-scale comparison of k2overall for HLA-A*01:01, HLA-A*30:01, HLA-A*29:02, HLA-A*02:01, HLA-A*68:02, HLA-A*03:01, HLA-A*11:01, HLA-A*24:02, HLA-B*08:01, and HLA-B*37:01 in the presence of hTAPBPR or chTAPBPR. The apparent rate constant kapp was determined by fitting the raw trace to a monoexponential association model. The extrapolation of the slope between kapp and the concentrations of TAPBPR orthologs determines the overall rate k2overall. NA indicates no k2overall (no peptide exchange) determined. Results of three technical replicates (means ± σ) are plotted.