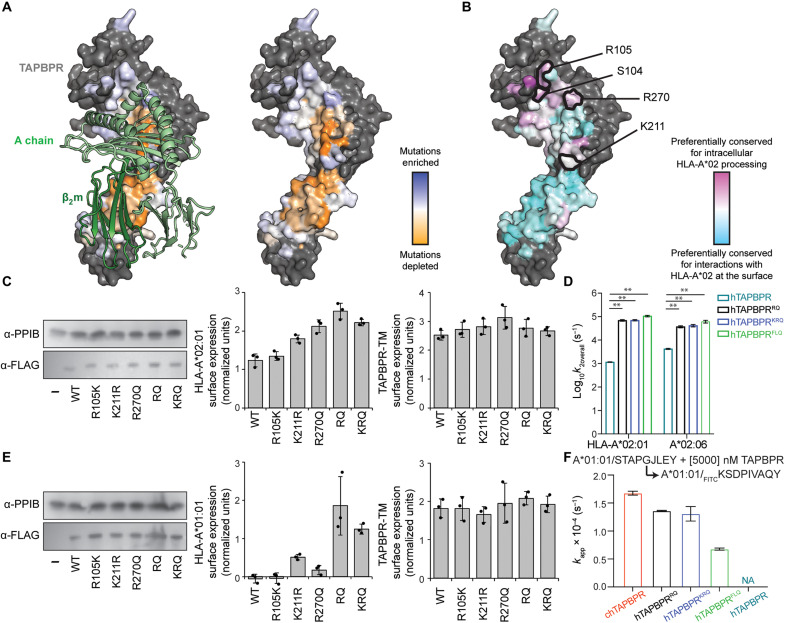
Fig. 6. Deep mutational scanning of TAPBPR surface enhances peptide exchange function for HLA-A*02:01 and HLA-A*01:01.
(A) Sequence conservation from deep mutagenesis is mapped to the surface of hTAPBPR (24) (PDB ID: 5WER, mouse H2-Dd/hβ2m shown as light and dark green ribbons at left). Mutationally depleted or tolerant residues are colored orange or white and blue. Nonmutated residues are gray. (B) Conservation scores from the deep mutational scan of hTAPBPR-TM versus hTAPBPR (50). Residues preferentially conserved for intracellular processing or surface interaction of HLA-A*02:01 are pink or cyan. Positions S104, R105, K211, and R270 are identified as more conserved for intracellular HLA-A*02:01 processing and are sites of gain-of-function mutations for promoting surface HLA-A*02:01 trafficking. (C) Immunoblots comparing total expression levels for TAPBPR mutants (α-FLAG) versus PP1B as a loading control (left). Surface expression of HLA-A*02:01 in the presence of the different TAPBPR-TM mutants (middle). Surface expression for the different TAPBPR-TM mutants (right). (D) Log-scale comparison of k2overall for HLA-A*02:01 and HLA-A*02:06 in the presence of hTAPBPR or mutants. The two-sample unequal variance Student’s t test was performed, P > 0.12 (ns), *P < 0.033, **P < 0.002, and ***P < 0.001. (E) Immunoblots comparing total expression levels for TAPBPR mutants (α-FLAG) versus PP1B as a loading control (left). Surface expression of HLA-A*01:01 in the presence of the different TAPBPR-TM mutants (middle). Surface expression for the different TAPBPR-TM mutants (right). (F) Comparison of kapp for fluorescent peptide binding to HLA-A*01:01 in the presence of 5 μM chTAPBPR, hTAPBPR mutants, and hTAPBPR. Results of three replicates (means ± σ) are plotted.