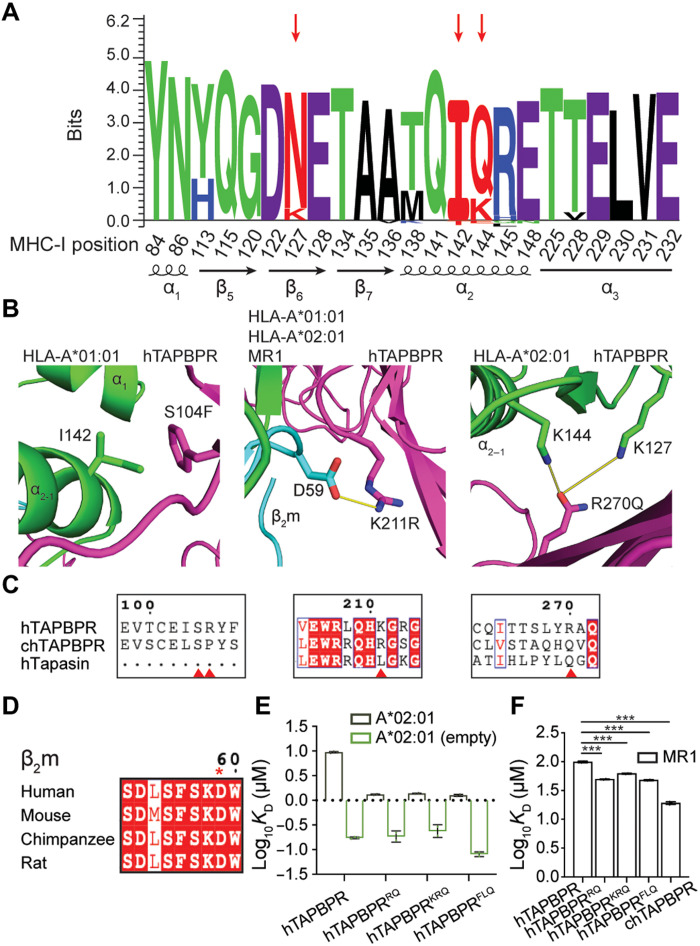
Fig. 7. Targeted mutagenesis of hTAPBPR surfaces can promote interactions with conserved or polymorphic epitopes on classical and nonclassical MHC-I molecules.
(A) Seq2logo (80) visualization of MHC-I residues that interact with TAPBPR calculated from the sequences of 75 different HLA allotypes. Sequence weighting used clustering, pseudo-count with a weight of 0, and Kullback-Leibler logotype. The percentage frequency of amino acids on a specific position higher than 10% is shown on the positive y axis and less than 10% amino acids on the negative y axis. (B) Structural models of HLA-A*01:01 or HLA-A*02:01 in complex with hTAPBPR generated by RosettaCM (72) (BAKER-ROBETTA server). Amino acids at positions 142 of HLA-A*01:01, 59 of hβ2m, and 127 and 144 of HLA-A*02:01 are indicated. (C) Sequence alignment of hTAPBPR, chTAPBPR, and human tapasin (hTapasin) near positions 104, 105, 211, and 270. (D) Sequence conservation patterns within the same region of β2m from the amino acid position 58 to 60 across four species. (E) Log-scale comparison of SPR determined KD values for peptide-loaded and empty HLA-A*02:01 interacting with hTAPBPR and TAPBPR mutants. (F) Log-scale comparison of SPR determined KD values for MR1 C262S interacting with hTAPBPR, TAPBPR mutants, and chTAPBPR. The two-sample unequal variance Student’s t test was performed, P > 0.12 (ns), *P < 0.033, **P < 0.002, and ***P < 0.001. Results of at least two technical replicates (means ± σ) are plotted.