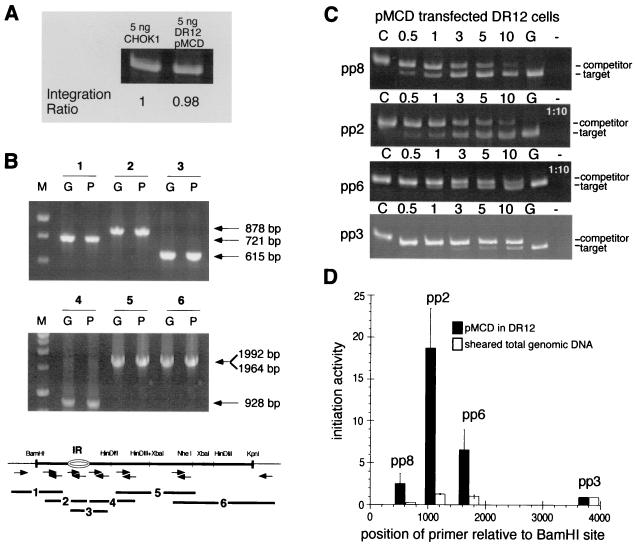
FIG. 3.
Initiation of DNA replication in the exogenous 5.8-kb ori-β fragment in DR12 cells. (A) The integrated exogenous DHFR ori-β fragment in 5 ng of DNA from uncloned pools of DR12 cells and the endogenous ori-β in 5 ng of CHOK1 DNA were amplified by PCR. The ratio of the amplification products in DR12 relative to those of endogenous ori-β in CHOK1 is indicated, and this ratio suggests that the copy number of exogenous ori-β fragments present in the pool of transfectants mimics that of the endogenous locus in CHOK1. (B) The structure of the integrated DHFR ori-β fragments in DR12 pools was determined by PCR amplification of either pMCD plasmid DNA (lanes P) or large single-stranded genomic DNA isolated from uncloned drug-resistant pools of pMCD-transfected DR12 cells (lanes G). Amplification reactions were performed with a panel of six primer sets spanning the 5.8-kb DHFR ori-β fragment and the flanking vector sequences (bottom). The resulting overlapping PCR amplification products, labeled 1 through 6, were analyzed by PAGE and ethidium bromide staining. M indicates a DNA size marker. The sizes of the resulting PCR amplification products are indicated. (C) PCR amplifications were performed with each of the four primer pairs and size-fractionated nascent DNA from asynchronous pMCD-transfected DR12 cells in the presence of a precalibrated amount of the corresponding competitor DNA. Amplification products were analyzed by PAGE and ethidium bromide staining. Control lanes: C, competitor template only; G, nascent genomic DNA template only; −, no template. Numbers above each lane represent the volume in microliters of nascent DNA added to the PCR mixture. Amplification reactions for pp2 and pp6 used a 1:10 dilution of the nascent DNA. (D) Amplification products generated with either nascent DNA from a pool of asynchronous pMCD-transfected DR12 cells (black bars) or sheared total genomic DNA from asynchronous pMCD-transfected DR12 cells (white bars) were quantitatively evaluated for seven independent transfection experiments. As a measure of initiation activity, the abundance of each target sequence in nascent genomic DNA was normalized to the abundance of pp3 target sequences in the corresponding experiment, which was set equal to 1 (see example in Table 2), and the average of the seven experiments is shown. Bars indicate the SEM.