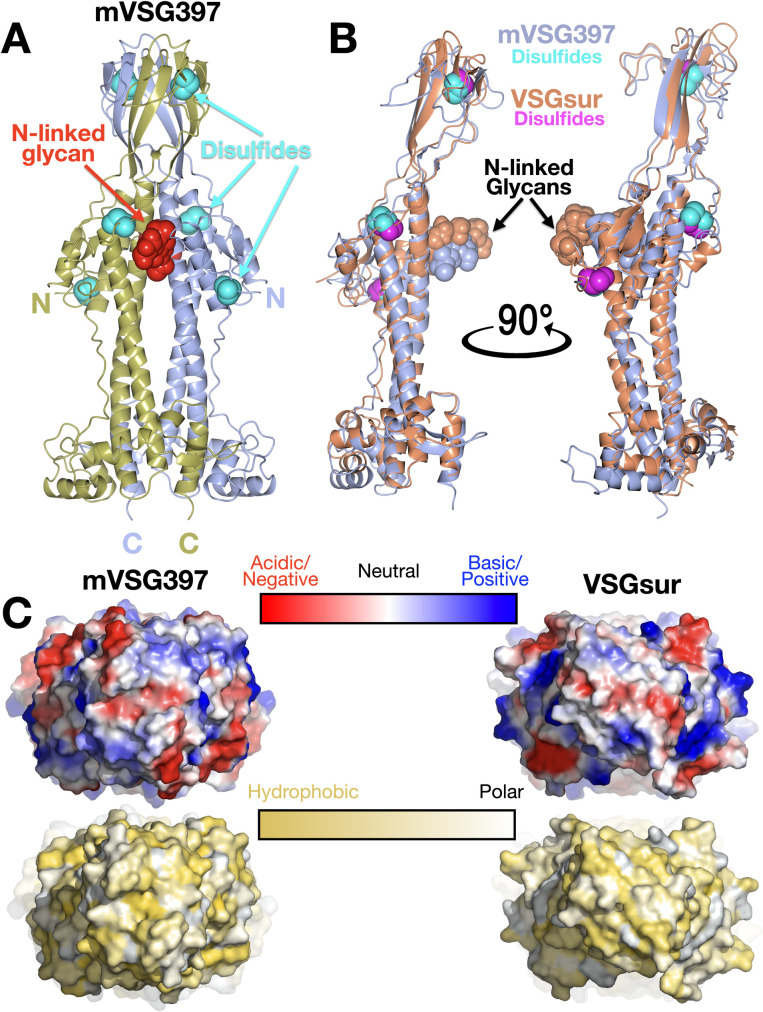
Fig 2. Crystal structure of mVSG397.
(A) Ribbon diagram of the (crystallographic) dimer of mVSG397, the individual monomers colored gold and light blue. The N-linked glycans are displayed as red space-filling atoms and disulfide bonds are shown in cyan. ‘N’ and ‘C’ indicate the N and C termini of each monomer. Structures drawn with CCP4mg [57] (B) Structural alignment of monomers of VSGsur (orange) and mVSG397 (light blue) with corresponding glycans and disulfides shown in space filling representation, the sugars colored the same as the protein to which they are linked, the disulfides in cyan (mVSG397) and magenta (VSGsur). Structures drawn with CCP4mg [57] (C) 90-degree rotation of the structures in (A) and (B) to view the “top” of the VSGs, rendered as molecular surfaces. Top row shows the surface colored by electrostatic potential (red indicating acidic/negatively charged, blue indicating basic/positively charged, and white neutral, all surfaces scaled identically). The bottom row is colored by the Eisenberg hydrophobicity scale [59], where yellow indicates hydrophobic and white polar. Molecular surfaces are generated by PyMOL [60].