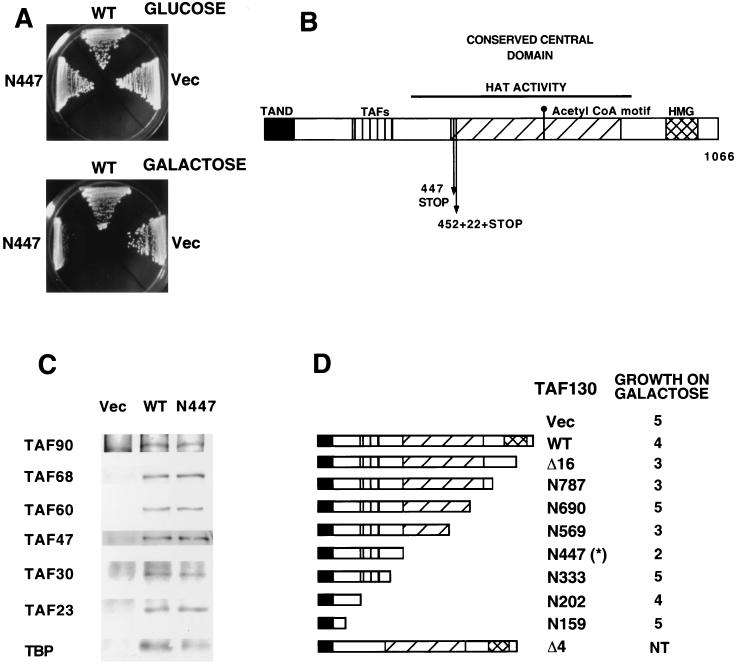
FIG. 1.
A dominant-negative TAF130 mutant that forms a normal TFIID complex. (A) Growth of yeast cells containing the plasmid vector or derivatives bearing wild-type TAF130 or the TAF130-N447 mutant expressed from the GAL1 promoter on plates containing glucose or galactose as the sole carbon source. (B) Location of dominant-negative mutations (vertical arrows) with respect to structural and functional features of TAF130. These features include a conserved central domain that has histone acetylase activity (31), a putative acetyl coenzyme A binding site (10), a putative HMG domain (44), an N-terminal inhibitory region (TAND) that strongly interacts with TBP (22, 29), an additional region interacting with TBP (2), and a region interacting with other TAFs (2). (C) TFIID complex formation. HA-tagged wild-type TAF130 or TAF130-N447 was immunoprecipitated from cell extracts with anti-HA antibodies, and components of the TFIID complex were detected by Western blotting with the indicated antibodies. (D) Dominant-negative effects due to overexpression of C-terminal deletions mutants of TAF130 (number indicates last residue of the protein). Growth phenotypes were determined on galactose medium and are defined in arbitrary units with a value of 5 indicating normal growth and a value of 0 indicating no growth. The structures of TAF130-Δ4 (lacks residues 208–303) and TAF130-Δ16 (lacks residues 913 to 1037) are also indicated. The asterisk denotes the N447 mutant isolated in the genetic screen.