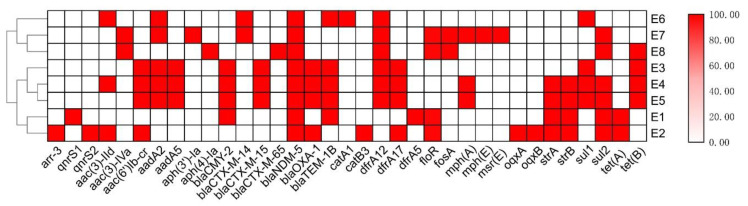
Figure 2.
Heatmap of antimicrobial resistance genes carried by E. coli isolates in this study. The horizontal axis represents the antimicrobial resistance genes, and the vertical axis represents the strain IDs. The red boxes represent the presence of the corresponding items among the sequenced isolates, and the white boxes represent their absence. The gradient identity bar indicates the percentage similarity of related genes. The similarity tree was constructed using agglomerative hierarchical clustering, with the degree of similarity between different clusters calculated by the average linkage method and the degree of similarity of different isolates (calculated using the Spearman’s rank correlation coefficient).The classes of antibiotics to which the genes confer resistance included aminoglycosides: strA, strB, aac(3)-IId, aac(6)-Ib-cr, aadA2, aadA5, aph3-Ia, aph(4)-Ia and aac(3)-IV; fosfomycin: fosA; fluoroquinolones: qnrS1 and qnrS2; macrolides: mph(A), mph(B), mph(E) and msr(E); chloramphenicol: catA1, catB3, and floR; rifampicin: arr-3; sulphonamide: sul1 and sul2; tetracycline: tet(A) and tet(B); trimethoprim: dfrA5, dfrA12 and dfrA17; extended-spectrum β-lactams: blaCMY-2, blaTEM-1D, blaOXA-1, blaCTX-M-14, blaCTX-M-15, and blaCTX-M-65; carbapenems: blaNDM-5.