Figure 1.
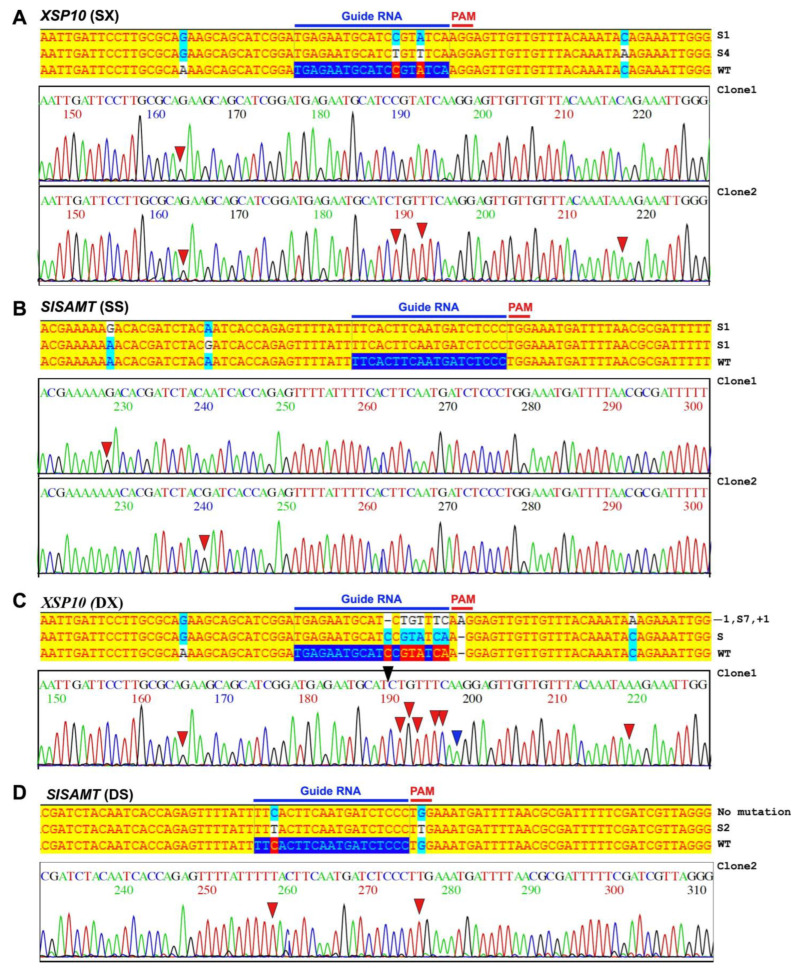
Evaluation of single-gene and dual-gene CRISPR editing events of XSP10 and SlSAMT through Sanger sequencing. (A) XSP10 single-gene editing (SX) events showing 1-bp (S1) and 4-bp (S4) substitution mutations in clone 1 and clone 2. Wild-type (WT) sequence of XSP10 used as a reference sequence. The mutations in nucleotides of each clone indicated as ‘−’ for deletion, ‘+’ for insertion, and ‘s’ for substitution. The bold blue arrows underlined 19-nucleotide sgRNA seed sequences along with PAM sites in the reference gene sequence. In the chromatogram, the red arrow indicates ‘s’, blue indicate ‘+’, and black ‘−’ for insertion. (B) SlSAMT single-gene editing (SS) events showing 1-bp (S1) substitution mutation in clone 1 and 2. (C) Dual-gene editing (DXS) of XSP10 (DX) showing 7-bp substitution (S7), 1-bp insertion (+1), and 1 bp-deletion (−1) observed in clone 1. (D) SlSAMT (DS) showing 2-bp substitution (S2) in clone 2 and no mutation in clone 1.