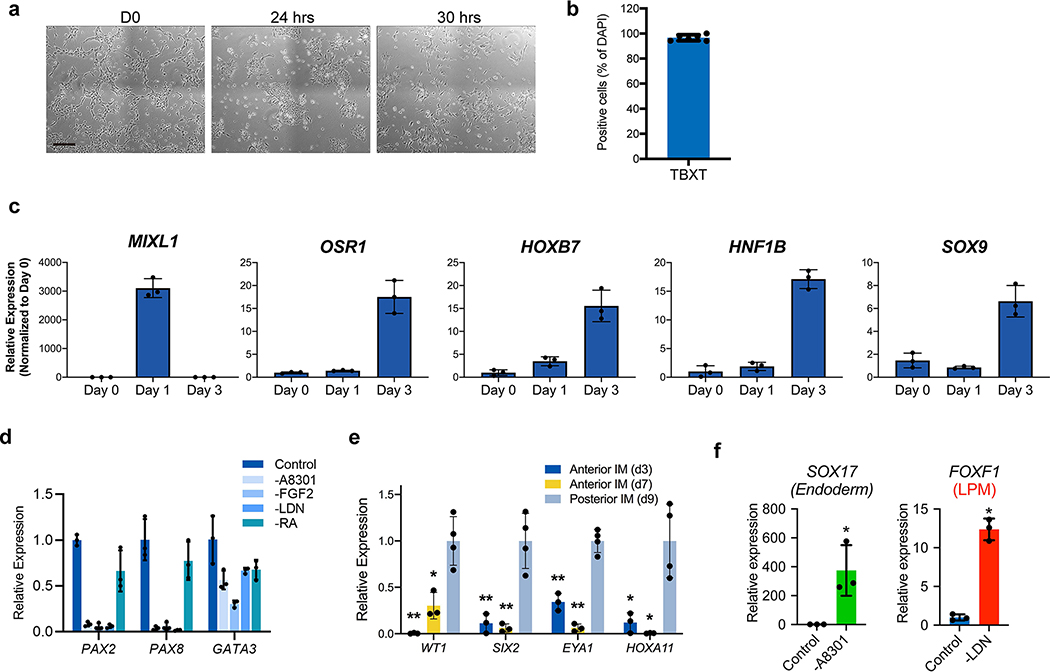
Extended Data Fig. 2. Efficient induction of pronephric IM cells at day 3.
a, Brightfield micrographs demonstrating appearance of undifferentiated hESCs at day 0, as well cultures after 24 and 30 hours exposure to primitive streak-inducing factors. At 24 hours, there was still significant colony-like morphology consistent with incomplete induction of mesendoderm cells, but after six additional hours the colonies were nearly completely dissociated into single, mesenchymal-like cells. b, Quantification of IF staining for TBXT (as shown in Fig. 1b) revealed >95% efficiency after 30 hours exposure. n=6 quantified fields from three independent replicates. c, Timecourse qPCR analysis corresponding to Figure 1c. The primitive streak marker MIXL1 was maximally expressed at day 1 and was subsequently quickly down-regulated, while IM markers OSR1, HOXB7, HNF1B, and SOX9 were increased by day 3. n=3 independent biological replicates per timepoint. d, Efficient specification and expression of pronephric IM genes at day 3 was dependent on the combinatorial effects of FGF2, RA, TGFβ inhibition (A83-01), and BMP inhibition (LDN193189) during days 1–3. n=3 independent biological replicates per condition. e, In the pronephric IM cultures at days 3 and 7, there was low level of expression of posterior IM markers (WT1, SIX2, EYA1, and HOXA11), which were derived from a differentiation protocol for inducing metanephric progenitor cells. *, p<0.05; **p<0.005; two-tailed Student’s t-test individually comparing day 3 and day 7 against the day 9 posterior IM samples; n=4 biological replicates, data representative of 2 independent experiments. Specifically p-values for comparisons using day 3 were 0.005, 0.006, 0.0005, and 0.017 for WT1, SIX2, EYA1, and HOXA11, respectively; at day 7, they were 0.007, 0.007, 0.0002, and 0.015. f, From days 1–3, the TGFβ inhibitor A83-01 was required for suppression of definitive endoderm (SOX17) fate, whereas BMP inhibition with LDN193189 inhibited formation of lateral plate mesoderm (FOXF1). *p<0.005; two-tailed Student’s t-test; n=3 biological replicates, data representative of 2 independent experiments. Scale bar 200 μm (a). Column and error bars represent mean and standard deviation, respectively.